Effective and rapid sequencing methods developed for proteins and nucleic acids have facilitated
the accumulation of a large volume of protein and DNA sequences.
The abundant sequence information cataloged in different databases promoted easy and rapid
elucidation of structure-function relationships in protein and DNA.
Development of similar sequencing strategies for linear polysaccharides in general and
HLGAGs in specific has been limited by their structural diversity
and the lack of information regarding template-based synthesis of these molecules.
There are 32 different disaccharide building blocks for HSGAGs, compared to 20 amino acids
for proteins and 4 nucleotides for DNA. Currently there are a few limited approaches
for sequencing these complex polysaccharides.
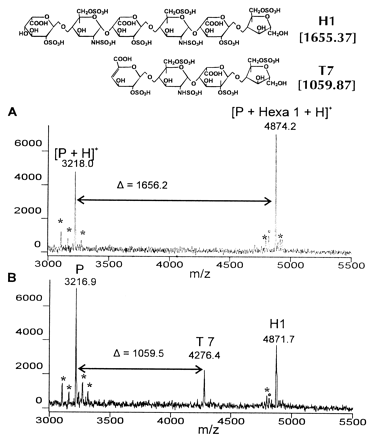
We have developed a rapid and effective sequencing strategy for HSGAGs that makes use powerful computational methods to manipulate experimental data obtained by chemical or enzymatic degradation of
HSGAGs, to obtain the final sequence of an unknown sample. In this approach we make use of the unique mass signatures of
HSGAG fragments, obtained from MALDI mass-spectrometry of the degradation products, as constraints to search a space of possible sequences for the correct sequence. The advantages of using MALDI mass spectrometry are the following:
 Using our rapid sequencing strategy and our HSGAG notation scheme, we aim to rapidly obtain sequences of HSGAGs that bind and activate different proteins in different physiological and pathophysiological processes. These sequences along with the already characterized HSGAG sequences (e.g. the antithrombin-binding pentasaccharide sequence) can be cataloged in the form of a database. An important factor that has hindered cataloging the existing HSGAG sequences is the unavailability of an elegant and easy-to-use representation scheme for the 32 disaccharide units. The use of single letter codes for protein and DNA has facilitated the development of sequence databases and rapid query systems to search sequences in the databases. We have developed a new coding scheme to represent the 32 disaccharide units not only to facilitate efficient cataloging of HSGAG sequences but also to improve the speed of the computational part of our sequencing strategy. The above methods will lead to rapid accumulation of HSGAG sequences and characterization of HSGAG sequences in the form of a database based on their properties including protein binding, biologically activity (which depends on protein binding). Important structure-function relationships of HSGAG sequences can be obtained from the HSGAG sequence database. |