Whitaker College of Health Science and Technology
Center for Environmental Health Sciences
The Center for Environmental Health Sciences (CEHS) pulls together the cross-disciplinary research and education efforts of some 27 members of the MIT faculty, plus five research staff scientists. In the past year, the center has undergone a change in leadership and research emphasis, with a program that applies a broad range of cutting edge technologies to the common goals of defining the impact of environmental agents on biological systems and identifying environmental causes of human disease. The center is funded by NIEHS and its associated research programs are funded through a variety of sources including NIGMS, NCI, DOE, NSF, ACS and DARPA. The many and varied research programs provide challenging interdisciplinary problems for postdocs, graduate and undergraduate students.
Simply stated, the mission of CEHS is to explore the biological effects of exposure to environmental agents, in order to understand and predict how such exposures affect human health.
The research activities in the center have been organized into six research cores, namely: signal transduction, mutation and cancer, free radical chemistry and biology, molecular, cell and tissue engineering for toxicology, environmental systems and health, computation and structure. A brief description of the goals of each research core are presented below.
The CEHS research activities are supported by four facilities cores that provide state of the art technologies for solving environmental health problems. The bioanalytical core facility and the accelerator mass spectrometry core provide central resources that provide expertise, training, and access to a wide variety of instrumentation, including (accelerator) mass spectrometry, liquid chromatography, and fluorescence spectroscopy. The Genomics, Proteomics and Bioinformatics Core Facility offers sophisticated DNA microarray analysis for transcriptional profiling as well as bioinformatics tools for the analysis of the full spectrum of cellular responses to environmental agents. Finally, the Molecular and Cellular Imaging Core provides state of the art cell and molecular imaging instrumentation, including 2-photon spectroscopy and laser scanning microscopy. Brief descriptions of each core facility are presented below.
The academic Biological Engineering Division allied with the Center for Environmental Health Sciences offers graduate education in molecular and systems toxicology and cross-disciplinary graduate opportunities in environmental health science and engineering with many departments in the Schools of Science and Engineering. Central to this educational effort, is an NIEHS-funded training grant. The molecular and systems toxicology core curriculum emphasizes integration of chemistry, molecular biology, and genetics with bioengineering approaches to understanding how organisms respond to environmental agents.
Research Cores
Mutation and Cancer Research Core
Directed by Professor John Essigmann, the objective of this research core is to use chemical, biochemical, genetic and whole animal approaches to understand the underlying mechanisms by which genetic change is induced following environmental insults. The expertise base of the core participants is very broad, including synthetic and analytical chemistry; mammalian, yeast and bacterial genetics; construction of transgenic animals; and expression array studies of mammalian and bacterial proteins and mRNAs induced following exposure to DNA damaging agents. Some probe the details of how specific gene products, most notably repair, replication and recombination proteins, remove DNA damage or allow cells to tolerate DNA damage. Others identify novel lesions formed by DNA damaging agents and determine the extent to which, and the manner in which, known enzymes process those lesions in vitro. Still others stratify the biological importance of individual lesions, both in terms of lesion toxicity and mutagenic potential. We study the sequence of events triggered by toxin and toxicant exposure and ending in apoptosis. We also use transgenic mice infected with various pathogens to understand how inflammation can trigger disease in intact animals.
Core members:
- John Essigmann (core director), professor, Biological Engineering Division and Chemistry
- Peter Dedon, associate professor, Biological Engineering Division
- Bevin Engelward, assistant professor, Biological Engineering Division
- Leona Samson, professor, Biological Engineering Division
- David Schauer, associate professor, Biological Engineering Division
- Steven Tannenbaum, professor and co-director, Biological Engineering Division
- Graham Walker, professor, Biology
Free Radical Chemistry and Biology Research Core
This core, directed by Professor Steve Tannenbaum, brings together CEHS members interested in understanding the chemical reactions of endogenous and environmental free radical species and the role of these processes in human disease. This highly interactive group is involved in interactive projects representing a program project grant and two RO1 grants. Two major areas are covered: the role of NO in mutagenisis and carcinogensis; the chemistry and biology of ionizing radiation and reactive oxygen species.
Core members:
- Steven Tannenbaum (core director), professor, Biological Engineering Division
- Jeffrey Coderre, professor, Department of Nuclear Engineering
- Peter Dedon, professor, Biological Engineering Division
- Bevin Engelward, professor, Biological Engineering Division
- John Essigmann, professor, Biological Engineering Division and Chemistry
- James Sherley, professor, Biological Engineering Division
- Gerald Wogan, professor emeritus, Biological Engineering Division
- Jacquelyn Yanch, professor, Department of Nuclear Engineering
Molecular, Cell and Tissue Engineering Research Core
Directed by Professor Linda Griffith, the mission of the MCTE core is development of new tools for analysis of toxicological phenomena across a hierarchy of levels—molecules, cells and tissues—via a synthesis of biology with engineering. The primary focus of this team is characterizing interactions of agents with eukaryotic cells and tissues, with an ultimate objective of predicting how to prevent or mitigate the effects of existing toxicants humans are exposed to, and how humans will respond to putative new toxicants, such as drugs, herbicides, etc. One emphasis of the team is building new model systems, such as 3D tissues that replicate the physiology of the liver capillary bed, new mice that allow quantitative study of specific recombination events, stem cell lines that enable analysis of asymmetric kinetics of cell division, and methods of screening the impact of toxicants on entire metabolic pathways. A complementary focus is development of new instrumentation methods to measure the properties of events occurring at several levels. Projects in this area include measurement of the mechanics of DNA under physiological conditions associated with damaging events and multiphoton microscopy to analyze apoptosis in 3D cultures, and rapid scanning multiphoton spectroscopy to assess rare recombination events in vivo. The team is also developing quantitative engineering models for that incorporate data from the measurements.
Core members:
- Linda Griffith (core director), professor, Biological Engineering Division
- Peter Dedon, associate professor, Biological Engineering Division
- Bevin Engelward, assistant professor, Biological Engineering Division
- James Sherley, assistant professor, Biological Engineering Division
- Peter So, associate professor, Mechanical Engineering
Research Core in Environmental Systems and Health
Directed by Professor Harry Hemond, the mission of this research core, is to understand, holistically, the relationships that link ecological processes and human health. Although this includes the now traditional "fate and transport" model (in which chemical releases are transported and modulated by processes in ecosystems, thus governing the extent of human exposure to the chemicals), advances of the past decade now mandate a broader view of environment/health linkages, in which genomics and ecology play an increasing role. Future advances will require better understanding of evolution, gene flow, and ecosystem processes along with progress in chemical and physical modeling and measurement. Gene flow, for example, can affect the distribution of pathogenicity, or the acquisition of antibiotic resistance or biodegradative capability in microbial communities. Ecosystem processes govern the nature of coexisting populations at scales from that of the gut to that of continents, with direct effects on humans at all scales.
This is an emerging field that cuts across traditional disciplines, and brings together researchers with expertise that includes but is not limited to environmental chemistry, ecology, microbiology, veterinary science, and environmental physics. A unifying theme is that of ecology in the broad sense; each project and researcher is involved with processes that occur in the natural environment, yet have implications for the well being of people. Increasingly, it can be seen that the well being of humans is inextricably interconnected with processes that may best be regarded as ecological, at all scales from planetary to cellular.
This area will become increasingly well defined, and recognized as critical to human health, in coming years. We want this core to contribute to the process. Examples of projects currently engaged by researchers in this core include: the environmental geochemistry of toxic metals, population dynamics of co-occurring pathogenic and non-pathogenic Vibrio species in natural waters, the ecology of the lower gut, the ecology and evolution of microorganisms in nature, and arsenic in drinking water in Bangladesh (a result of a tradeoff between chemical toxins and environmentally transported pathogens).
Core members:
- Harry Hemond (core director), professor, Civil and Environmental Engineering
- Sallie Chisholm, professor, Civil and Environmental Engineering
- James Fox, professor, Biological Engineering Division
- Charles Harvey, professor, Civil and Environmental Engineering
- Heidi Nepf, associate professor, Civil and Environmental Engineering
- Martin Polz, assistant professor, Civil and Environmental Engineering
- David Schauer, associate professor, Biological Engineering Division
Signal Transduction Research Core
Directed by Professor Douglas Lauffenburger, the cell signaling core comprises a half dozen scientists and engineers undertaking collaborative projects with the goal of developing quantitative, integrative systems understanding of cell-cell communication and intracellular signal transduction.
Substantial collaborative projects exist as connecting "edges" between many of these "vertices", some of which are being pursued as a major multi-investigator DARPA-funded program in Cell Decision Processes aimed at deciphering the "information flow" governing death-versus-survival decisions in human blood and tissue cells confronted simultaneously by death-promoting and survival-promoting soluble factors. Among this class of projects are a Sorger-Lauffenburger-Tannenbaum collaboration on kinetic modeling of TNFa-, EGF-, and Insulin-activated networks regulating caspase-mediated apoptosis processes and a Yaffe-Lauffenburger-Sorger collaboration on high-throughput quantitative protein kinase activity assays for generating dynamic data for vector state-space analysis of cue/signal and signal/response relationships. A nascent Lauffenburger-Samson collaboration extending the latter kind of systems approach to DNA damage activated death/survival decisions, and an analogous Yaffe-Sorger-Lauffenburger effort toward analyzing decision pathways activated by chromosomal segregation defects, are being built upon this foundation.
All of these collaborative investigations manifest the quantitative systems perspective catalyzed by our MIT biology/engineering fusion, and leverage funding support from other government agencies (including NIGMS, NCI, NSF, DARPA, and Army) as well as biotechnology and pharmaceutical companies.
Core members:
- Douglas Lauffenburger (core director), professor, Chemical Engineering and Biological Engineering, co-director Biological Engineering Division
- Ram Sasisekharan, associate professor, Biological Engineering Division
- Leona Samson, professor, Biological Engineering Division
- Peter Sorger, associate professor, Biology Department
- Steven Tannenbaum, professor, Biological Engineering, co-director Biological Engineering Division
- Michael Yaffe, assistant professor, Biology Department
Computation and Structure Research Core
The MIT Center for Environmental Health Sciences research core in computation and structure, directed by Professor Bruce Tidor, consists of both structural biologists and cell biologists with a wide range of expertise and areas of research interest. The unifying theme that brings this research core together is the development and use of computational tools to interpret and predict molecular structures and cellular behavior in response to exposure to environmental agents.
This newly formed research core already has a number of strong interactions and is expected to grow new links internally, with other research cores in the center, and with facilities cores of the center. Ellenberger's structural studies of DNA repair proteins involve extensive interactions with the mutation and cancer research core and the Free Radical Chemistry and Biology Research Core. Lauffenburger is director of the signal transduction research core and Sorger is co-director of the genomics, proteomics, and bioinformatics core facility. Tidor, Sorger, Lauffenburger, and Samson are on the executive committee of a new initiative in computational and systems biology at MIT, an umbrella program under development that leverages the expertise of a broad cross-section of the campus from different departments to foster research and education in this new and vigorous area. In general terms, joint students exist between members of the computation and structure core and almost every other research core in the center, while much of the computational and data modeling work will rely on data generated in other research cores, particularly so with the signal transduction research core currently. It is easy, however, to see the strengthening of other links facilitated through the center.
A significant portion of the computational and data modeling performed in this research core will make use of the resources of the genomics, proteomics, and bioinformatics core facility. Of particular importance here are Tidor's development of computational methods for informatics, Sasisekharan's glycoinformatics, Sorger's studies of cellular decision-making and Lauffenburger's cellular modeling studies. The work of structural biologists will benefit greatly from the bioanalytical core facility, with emphasis on the chemical analyses required for the studies performed by Ellenberger, Drennan and Sasisekharan. Finally, Sorger's imaging work will make use of the molecular and cellular imaging core facility.
Core members:
- Bruce Tidor (core director), associate professor, Biological Engineering Division and Electrical Engineering and Computer Science
- Catherine Drennan, assistant professor, Chemistry
- Tom Ellenberger, professor, Harvard School of Public Health
- Doug Lauffenburger, professor, Biological Engineering Division
- Ram Sasisekharan, associate professor, Biological Engineering Division
- Peter Sorger, associate professor, Biology
Core Facilities
Bioanalytical Core Facility
Co-Directors: Dr. Pete Wishnok and Dr. Koli Taghizadeh
Researchers who need extensive use of dedicated instruments will generally have these in their own laboratories. Situations often arise, however, where additional equipment is needed on a temporary basis, or where occasional use of a specialized instrument such as a mass spectrometer can't be justified. In other cases, researchers may need expert assistance in method development or experimental design. The bioanalytical core laboratories maintain an extensive collection of up to date major instruments, along with skilled and experienced scientists, to fulfill these needs.
Dedicated equipment (in Building 16):
- Agilent 5973 Benchtop GC-MS with electron ionization and positive and negative-ion chemical ionization
- HP 5973 Benchtop GC-MS with electron ionization
- HP 5973 Benchtop GC-MS with electron ionization
- Sciex API I LC-MS with electrospray and APCI ion sources
- Agilent LC-MSD single quadrupole electrospray system
- HP 1100 binary pumping system with diode-array, variable wavelength UV, and fluorescence detectors
- HP 1090 binary pumping system with diode-array and fluorescence detectors
- ISCO precision ternary syringe-pump system with diode array and variable wavelength UV detectors
- Perkin-Elmer preparative-scale LC system
Other Equipment:
- Fourier Transform Infrared Spectrophotometer
Equipment available ad hoc via the Biological Engineering Division Mass Spectrometry Laboratory (in Building 56):
- Agilent 1100 Capillary MSD ion-trap mass spectrometer with electrospray, photoionization, nanoelectorspray, and atmospheric-pressure chemical ionization sources
- Agilent 1100 LC MSD single-quadrupole mass spectrometer with precision mass-based fraction collector. All the ion-sources listed above for the MDS Trap are compatible with this instrument.
- Applied Biosystems API 3000 tandem quadrupole mass spectrometer with electrospray, atmospheric pressure chemical ionization, and nanoelectrospray ion sources and a dedicated HP 1100 binary pumping system with diode-array detector
- Perceptive Biosystems Voyager Elite DE MALDI-TOF mass spectrometer with delayed extraction and reflectronOther equipment includes several free-standing HP 1100 binary pumping systems with a variety of interchangeable detectors and modules including diode-array detectors, autosamplers, thermostatted column compartments, and a fluorescence detector; an automated desalting/column-switching system that can be used with any of the LC/MS systems; free-standing UV-vis spectrophotometer; SpeedVacs, etc. The lab contains a permanent facility for custom packing of capillary HPLC columns.
Equipment in each laboratory is connected to a local network for printing, data transfer and storage, and Internet access. As noted below, we are in the process of developing an inter-laboratory network.
The bioanalytical core is directed by the internationally renowned mass spectrometrist Dr. John S. Wishnok and co-directed by Dr. Koli Taghizadeh. Elaine Plummer, an experienced research specialist, is funded full time through CEHS. Joseph Glogowski, a technical specialist, is available ad hoc for software and hardware consulting.
The bioanalytical core will be used regularly by at least five of the research cores, i.e., signal transduction; radical chemistry and biology; mutation and cancer; molecular, cell, and tissue engineering, and environmental systems and health research cores. In addition, there will be developmental interactions with the accelerator mass spectrometry, the molecular and cellular imaging, and the genomics, proteomics, and bionformatics core facilities.
The field of bioanalytical chemistry—especially in mass spectrometry—is in a period of extremely rapid development. We expect in the near future to acquire at least one high performance mass spectrometer, e.g., a quadrupole time of flight or a tandem quadrupole/linear ion-trap. The dedicated CEHS laboratories, including offices for Dr. Taghizadeh and Ms. Plummer, will move to new space within a year or so. To facilitate data exchange and communication, a web-based network is being developed by Mr. Glogowski for our current facilities; this should be operational by the time of the move, and should include secure but accessible data storage and real-time video conferencing.
Accelerator Mass Spectrometry Core Facility
Directed by Dr. Paul Skipper, the accelerator mass spectrometry (AMS) core provides ultrasensitive detection and quantitation of biomedical and other organic samples that have been isotopically labeled with 14C or tritium. Research that involves such samples is anticipated to cover a broad range of interests, having in common the need to detect extremely low amounts of isotope deliberately introduced into the system of interest by the investigator. The AMS instrument is central to this core since there are very few available worldwide and most are dedicated to non-biomedical applications. The AMS also provides a unique service and expertise in integrating AMS detection with conventional (GC, HPLC, multi-well plate) bioanalytical instrumentation.
Equipment
The principal instrumentation is a compact, low energy AMS that was designed and constructed by Newton Scientific, Inc., and installed and brought on line at MIT. From the beginning, this instrument was intended to be operated as a bioanalytical instrument, with connections to chromatographic and other sample purification and separation systems. This operational mode distinguishes it from other AMS instruments, which accept samples only in isolated solid—or, rarely, gaseous—form. The AMS has been operational for approximately two years and undergoes a continuous process of evolution and upgrading. At present, the detection limit for 14C is 10-18 mole in samples with 14C:12C isotope ratio of 10-10.
Major commercially available equipment includes an HP5890 gas chromatograph, which will soon be equipped with a Thermal Desorption System for injecting trapped gas samples, and an Agilent 1100 capillary HPLC. Both of these have been interfaced to the AMS as described below.
The GC column output is directed into a CuO reactor that oxidizes samples as they emerge from the column to produce CO2 from the sample carbon. CO2 is then transported in a carrier gas stream into the AMS ion source for analysis. This interface is based on previously described designs used for isotope ratio gas chromatography-mass spectrometry. It incorporates unique features needed for successful operation with AMS.
For introducing non-volatile samples, we have developed an interface based on laser-induced combustion for rapid conversion of organic carbon to CO2 and subsequent transport of the CO2 formed into the ion source of the AMS, thereby eliminating the conventional graphitization process used to produce solid samples. Sample is applied to a layer of CuO catalyst deposited in a refractory support. Volatile solvent is removed by evaporation. The catalyst plate is then translated through a reaction chamber in which it is irradiated by an infrared laser beam. Localized heating of the catalyst layer by the laser induces combustion of sample carbon to CO2. A constant flow of He removes the CO2 directly from the site of formation and transports it to the AMS ion source. Individual samples are irradiated in sequence for AMS analysis independently of the other samples present. Applicability of the same system for analysis of HPLC has also been demonstrated. Instead of applying individual samples at different locations on the catalyst layer, HPLC eluent is applied continuously to the bed as it moves past a deposition point at a constant rate. The overall process preserves the essential features of the chromatogram.
A prototype interface for tritium-labeled samples has also been designed, fabricated, and tested. This interface is based on pyrolysis, rather than combustion chemistry, and accepts solution samples without desolvation. Because it is designed for very small samples, its ultimate applicability is expected to be to high-throughput, micro array-based sample processing.
Since this core is being newly established, in the immediate future it will focus on the activities for the various Center investigators. Longer term, there are other directions already being pursued. We currently have a grant proposal under review that would fund the design and construction of a new, tritium-only, combination AMS instrument/high throughput interface. The design philosophy behind this combination is to produce an instrument that is capable of high resolution, rapid interrogation of tissues and sample arrays derived from experiments utilizing lower-cost and more readily accessible tritium-labeled compounds. There will also be a continuing program of hardware and software upgrades to the existing AMS instrument and interfaces. There is an interest on the part of Professor Tannenbaum to expand his research into single-cell metabolism studies by detection of metabolic 14CO2 in microfluidic devices.
Genomics, Proteomics and Bioinformatics Core Facility
The MIT Center for Environmental Health Sciences research core in Genomics, Proteomics and Bioinformatics is co-directed by Professor Peter Sorger and Dr. Rebecca Fry and is integrated into the MIT BioMicro Center. The BioMicro Center was established in 2000 to acquire and operate robotic instrumentation for microarraying and computer systems for bioinformatics, and to provide core services in these areas. It is a joint endeavor of the Center for Environmental Health Sciences, Department of Biology, Center for Cancer Research and Biological Engineering Division. The BioMicro Center aims to provide an integrated facility for microarray fabrication, microarray analysis, database storage, data mining and data modeling.
For users to derive accurate and meaningful microarray data in a timely fashion, it is essential that fabrication and computational services be supported with a high standard of excellence. This in turn requires a professional staff of research scientists and strong oversight committees. The BioMicro Center currently has a staff of seven, three of whom are devoted to array technologies and four to information technology. Day to day operations of the BioMicro Center are supervised by Peter Sorger, director, and center policies set by an executive committee comprising of Leona Samson, Doug Lauffenburger, Bob Sauer and Tyler Jacks.
The BioMicro Center is currently being relocated to newly refurbished labs on the third floor of the Koch Biology Building. Computing clusters are also being set up in Building 56 and the Cancer Center. The BioMicro Center aims to support genomics, informatics and microarray research in six key areas:
- Integrated support for Affymetrix GeneChips and Spotted Microarrays—The BioMicro Center supports the routine production and custom fabrication of spotted DNA microarrays based on oligos and cDNA and will also support Affymetrix GeneChips. It seems almost certain that both spotted arrays and GeneChips will be important for expression profiling experiments over the next few years. Spotted microarrays have the advantage of lower cost and greater flexibility. GeneChips are becoming increasingly important when whole-genome coverage is needed. Currently, human, mouse and yeast spotted arrays are available from the core for a cost of $75–$150 each.
- Strong three tier architecture for data storage and processing—Expression profiling can be performed on a small scale using spreadsheets and desktop applications. As the amount of data increases however, it is essential to use databases. The BioMicro Center has installed several three tier client/server systems based on open source and commercial database management systems (DBMS). These systems will be strengthened and extended to achieve integrated support for GeneChips and spotted microarrays.
- Network of managed desktop computers for data analysis and mining—The BioMicro Center has licensed commercial software including the Affymetrix MAS-DMT suite and Spotfire Decision Site for basic analysis of microarray data. The number of licenses will be increased and open-source tools will be integrated into the three tier BioMicro IT system. Client software will then be deployed on a network of managed desktop computers based on the MIT Win-Athena and Linux-Athena environments. The desktop network will provide a distributed system of analysis software tightly integrated to a high-reliability data storage infrastructure.
- Training and educational program in commercial and open-source bioinformatics software—A program of professional training seminars for commercial software will be strengthened and supplemented with seminars taught in-house for open source software.
- Technical assistance with programming, DBMS administration and data analysis—Additional staff is currently being hired to provide a strong capability in programming and database customization. Bioinformatics applications that currently operate separately from each other will be integrated.
- Quality control, error modeling and new technologies introduction—Rebecca Fry is overseeing the development of data models and quality control procedures for DNA microarray analysis. Error models will be integrated into a comprehensive Bayesian informatics chain. The introduction of new devices and computational methods will be accelerated through close collaborations between biologists, engineers and computer scientists throughout MIT.
In the area of microarraying and bioinformatics, the immediate goal of the BioMicro Center is to fully implement a database driven workflow for the analysis of spotted and Affymetrix gene arrays. This will include completion of our Oracle-based databases for array data, the integration of desktop software with these databases, and the installation of managed clusters of desktop computers for running software. Substantial completion is expected by fall 2002.
In the area of high performance computing, the BioMicro Center is currently installing a 64-processor Beowulf cluster computer and has funds to expand to 128 processors. This system will be complemented by a new 6TB-class data storage system and a gigabit ethernet network. The Beowulf system is on order and the Bio-SAN network is currently being installed. Support for high performance cluster-based computing will be in place by late summer 2002 and is made possible by a grant from the NSF.
In the longer term, the BioMicro Center intends to link its fabrication facilities with the MIT Microsystems lab and the rapid prototyping lab in the Media Lab. This will assist in the development of new devices combining proteins and DNA with microfabricated devices.
The Molecular and Cellular Imaging Core Facility
The Molecular and Cellular Imaging Core, directed by Dr. Elena Gostjeva, will be a crucial resource for CEHS researchers. State of the art molecular and cellular imaging is absolutely required for today's biological research, and research in the Environmental Health Sciences is no exception. In particular the ability to measure the influence of environmental agents on genetic, biochemical and biological processes in cells—whether in single celled cultures or multicellular tissues —is crucial for much of the research carried out by CEHS faculty. One goal of the core is to provide molecular cytological analysis that includes the best solutions in visualization and measurement of microscopically detectable objects within diverse biological systems. The development of automated programs that would speed up scanning, counting and measurements of specific biological targets will be pursued. The overall objectives of this core are: to provide training in sample preparation, and training in the efficient use of the available instrumentation, for all CEHS members and their students, postdocs and staff; and to develop new imaging methods and analytical tools.
Sophisticated microscopes and sensitive imaging instrumentation are required for monitoring the presence and location of proteins in cells and tissues (using immunofluoresence or fluorescently tagged proteins), for monitoring the induction of damage to chromosomes (cytogenetics), for monitoring the induction of cell death by apoptosis or necrosis, for monitoring the induction of cell cycle checkpoints, and for monitoring pathological changes in cells and tissues. All of these endpoints can be altered or triggered in response to environmental agents. The Molecular and Cellular Imaging Core is fairly well equipped for measuring each of these endpoints. The currently available instruments, and those that we plan to incorporate in the future are detailed below.
Current Equipment:
- Nikon Eclipse E800 fluorescence microscope, comprising four Nikon objectives (Plan Fluor 10x fluorescence; Plan Apo 60x achromatic; Plan Apo 100x achromatic), a 100W Hg Epifluorescence Arclamp, an Optronics cooled color CCD camerawith DEI-750 digital image processing electronics, 4 filter blocks, and an Apple Power Macintosh 8600/200 computer with Scion Corp. CG-7 Frame Grabber video capture card
- Nikon Labophot fluorescence phase-contrast microscope, comprising 7 Nikon objectives (10x, 20x, 40x, and 100x phase-contrast; 40x and 100x fluorescence; Plan Apo 100x achromatic), a 6-position phase ring with darkfield setting, 5 filter blocks, 100W Hg Epifluorescence Arclamp; Nikon 35mm camera and Nikon AFX exposure control apparatus
- Zeiss fluorescence phase-contrast microscope, comprising 5 Zeiss objectives (10x, NEOFLUAR 25x, 40x, and 100x fluorescence phase-contrast; and Planapo 63x achromatic), a 100W Hg Epifluorescence Arclamp, a 35mm camera and a Zeiss MC63A exposure control apparatus
- CompuCyte LSC Laser Scanning Cytometer, comprising a HeNe laser, 3 filter/photomultiplier tube units, cytometer, an Olympus BX50 fluorescence microscope with proprietary modifications, 3 UPlanFluor fluorescence objectives (10x, 20x, 40x) a black-and-white CCD camera with CRT display, a color 3-CCD camera with CRT display, 3 filter blocks, an HP Vectra VL computer with proprietary data acquisition and image capture components
- Zeiss phase-contrast microscope, comprising 4 Zeiss objectives (10x, 25x, NEOFLUAR 40x fluorescence phase-contrast, 100x), 4-position phase ring with darkfield setting
- Bausch & Lomb dissecting microscope
- Zeiss Axioskop 2 MOT fluorescence phase-contrast microscope, comprising 3 Zeiss objectives (Plan NEOFLUAR 10x and 20x fluorescence, Plan Apo 100x achromatic); phase-contrast, darkfield, and DIC optics; precision motorized stage and focus controls with MCX-2 high resolution positioning controller; 3 filter blocks; 100W Hg epifluorescence arclamp; and a Zeiss AxioCam color, ultra-high resolution (3900 x 3090 pixels) digital camera connected to a PC workstation
Future purchases that are currently under discussion include a Molecular Dynamics Typhoon Phosphorimaging instrument for detection, imaging and quantitation of radioactive, chemiluminescent and fluorescent molecules. In addition, CEHS members have requested that instrumentation for laser capture microscopy (LCM) be added to this core facility and, should there be enough demand for such a capability, we will act upon this request.
The ultimate goal of the molecular and cellular imaging core is to enable researchers in the labs of the CEHS faculty to apply cutting edge microscopy and sophisticated imaging in their quest to determine how environmental agents perturb biological systems. This type of analysis, i.e., at the tissue, cellular and subcellular levels, dovetails perfectly with the kind of information obtained by the bioanalytical core facility and the genomics, proteomics and bioinformatics core facility. Dr. Elena Gostjeva has fifteen years of experience in light microscopy and cytogenetic analyses of various different types of chromosomes. Her extensive experience with state of the art microscopes and advanced digital imaging methods prepares her well to be director of this core. Use of the aforementioned instruments will be overseen by Dr. Elena Gostajeva, and she will train lab members of the CEHS faculty in the methods required for preparation and analysis of biological materials for immunohistochemistry, cytogenetics, cytotoxicity and other related microscopy and imaging methods. Unfortunately she is only able to commit 30 percent of her time to this endeavor, and we therefore plan to recruit a co-director for this core. Our plan is to appoint a co-director who will specifically interface between the CEHS molecular and cellular imaging core and a number of other imaging cores at MIT that also offer sophisticated light microscopy facilities. In this way we hope that the imaging cores on campus will co-ordinate their purchases and avoid needless duplication.
For future core capabilities we plan to develop the following:
- automated labeling/counting of apoptotic cells in human cell cultures and tissues
- measurement of DNA damage through yeast cells comet assay
- automated program of 2D images deconvolution in a very small size objects (on a range of about 0.1 um, 4x105–1x104 DNA base pairs length) using yeast mitotic chromosomes as a model
- standardization of the image analysis with regard to measurement of eukariotic cell nuclei sizes and DNA contents (internal standard for visual image cytometry approach)
With the assistance of the Zeiss Company we also hope to develop the following:
- to establish the best resolution in microscope phase contrast imaging of non-stained cells (lenses versus image deconvolution programs): excellent for the study of morphology and architecture of cells, nuclei and chromosome structures; the application area: apoptotic cells, genomic instability; structural chromosome rearrangements.
- to establish the best imaging resolution of multiple stained cells, FISH stained chromosomes and chromosome structures, anti-body stained proteins; possible application area: study of the genetic apparatus damage at the level of large-scale chromatin.
- To probe the PALM Laser-MicroBeam system (micro-tweezers) to catch and move a single cell, a cluster of cells from the microscopic slide into a tube for molecular analysis; possible application area: stem cell specific mutations; cell-specific gene expression analysis in living tissues.
These methods are thought to be of importance as they might fill the gap between studies at molecular and cytogenetic levels in understanding how environmental agents interact with the living cells on a pathway to human disease.
Community Outreach and Education Program
The Community Outreach and Education Program (COEP) program is directed by Professor Heidi M. Nepf, who collaborates with Amy Fitzgerald, director, Edgerton Outreach Program, MIT. In a recent speech entitled, "Science as Patriotism," NSF Director Dr. Rita Colwell stressed both, "the primary importance of a scientifically literate citizenry," as well as, "the responsibility of the science and engineering community to meet that goal." Dr. Colwell went on to give this recent example of how misinformation can breed chaos and hysteria. During the anthrax scare many public officials and private citizens believed that the disease was contagious—a misconception that exacerbated public stress. Perhaps more compelling are the daily decisions faced by citizens, from personal health care to community development, which require the interpretation of scientific information. Finally, the level of scientific literacy of high school students greatly influences the pipeline of young talent entering the scientific and engineering workforce upon which our technological society depends. The goal of the MIT COEP is to promote of scientific literacy with a variety of projects targeted to students in grade four through undergraduate. In addition, the MIT COEP mentors young scientists on the mechanisms and importance of educational outreach by supporting the participation of undergraduate and graduate students in the development and implementation of outreach activities.
Highlights From COEP Activities
Collaboration with MIT's Edgerton Center
 Grungy Groundwater in action. Cambridge Public School students learn about groundwater transport and pollution using models they construct themselves. |
Through hands on activities, the Edgerton Center invites students to be scientists for a day. This past year the center hosted 2000 students from public schools and community groups. To take advantage of its infrastructure, COEP bought into the Edgerton Program, beginning in 2001, by supporting one quarter of the program director's salary. Working with the director, Amy Fitzgerald, and with guidance from the Cambridge Public School teachers we developed a new activity, Grungy Groundwater, that challenges students to discover how water and pollutants move underground, and how pollutants can impact drinking supplies. Students first explore how fluids travel through different soil types. Then, students build their own models of the underground using different soil types. The students use their models to discover how buried and surface contamination enter and travel through the subsurface. The session ends with a discussion of how a community might respond after discovering contamination in their drinking wells.
During the past year the activity was tested and edited by a group of Cambridge Public School (CPS) teachers and students. The activity will be a formal offering of the center starting next school term (fall 2002), and we expect about 500 students per year will take part. In addition, with cooperation from the CPS Science Coordinator, Dr. Melanie Barron, we hope to integrate this activity into the pollution and ecosystem health unit of every 5th grade class in the CPS system. A video of the Grungy Groundwater Model in action is available at http://web.mit.edu/edgerton/outreach/ACT_GAG.html. With the successful launch of Grungy Groundwater, we have just begun the development phase of a second activity.
More information about the collaboration with MIT's Edgerton Center can be found online at http://web.mit.edu/edgerton/outreach/out.html.
Video and Curriculum Development
Working with educational consultant Dr. Francesca Casella, COEP created a curriculum package on groundwater pollution and the superfund program. The video walks students through the investigation of a contaminated site and the process by which a site is added to the National Priorities List. A curriculum guide outlines supporting activities such as hands-on experiments, web-based learning exercises, and suggestions for library and community research. For example, students are asked to query environmental databases to identify existing superfund sites in their community and to research major pollutant inputs in their area and their potential health hazards. To help teachers integrate this package into the curriculum, the guide contains a chart mapping each activity to the National Science Education Standards. Initially the package will be disseminated through the NIEHS COEP Resource Center, directed by Karalyn Colopy, and through the Teacher as Scholars Program (see below). Portions of the video have already been adopted by the Advanced Technology Environmental Education Center (ATEEC) for use in a new curriculum for junior college students.
Teachers as Scholars Program
Teachers as Scholars is a K–12 teacher education program run jointly by Harvard and MIT. Selected teachers are invited to campus for seminars led by university faculty. Professors Culligan and Nepf host an annual a three day course, "Pollutant Transport in Natural Water Systems," that includes hands on laboratory activities. Teachers are encouraged to incorporate aspects of the seminar into their own curriculum. COEP facilitates this by providing advice, supplies and technical support to recreate all or part of the experiments for individual schools. For example, we adapted one of the laboratory exercises for Rebecca Green, a 7th grade teacher from Belmont Public Schools. We also developed an original water sampling activity for Jenn Morrell, a 6th grade teacher from Cambridge Public Schools. More information about the Teachers as Scholars Program can be found online at http://www.teachersasscholars.org/.
Pilot Program
The Pilot Program is being reorganized, and we have currently a request for proposal advertised to encourage faculty across the Institute to submit a proposal. We expect to fund five proposals in the coming year.
More information about the Center for Environmental Health Sciences can be found online at http://web.mit.edu/cehs/.
Clinical Research Center
The Clinical Research Center (CRC) was established in 1964, with grant support from the National Institutes of Health (NIH), to provide a facility in which MIT investigators and their collaborators could apply the Institute's expertise in basic biochemical and biophysical mechanisms to the analysis of normal and pathologic processes in humans. MIT's CRC was the first federally supported clinical research center located in a university and not within a hospital, and remains one of only two or three such centers. It was anticipated that in spite of its university venue, a large number of qualified physicians and clinical scientists from MIT's faculty and staff would utilize the CRC to study normal volunteers, or patients with chronic diseases.
Scientists and physicians authorized to carry out research protocols using the CRC's facilities include professors, research scientists who work exclusively at MIT, and those with primary appointments in local medical institutions whose research interests overlap extensively with those of MIT investigators. Research protocols must be approved by the MIT Committee on the Use of Humans as Experimental Subjects (COUHES) and the CRC Advisory Committee before they can be implemented. The CRC Advisory Committee, chaired by Dr. Daniel Shannon, professor of pediatrics at the Harvard Medical School and professor of health sciences at the Harvard/MIT Division of Health Sciences and Technology, consists of ten voting members plus nine non-voting members from the CRC's program and operating staffs. The committee has reported to the principal investigator of the CRC's NIH Grant, Martha Gray, professor and co-director of Harvard/MIT Division of Health Sciences and Technology (HST). With the CRC's administrative merger with the Massachusetts General Hospital's CRC, it now reports (for NIH grant purposes) to Dr. James Mongan, principal investigator of the joint NIH grant. It meets bimonthly to evaluate protocols for their scientific quality, experimental design, ultimate statistical validity and potential risk to human subjects. The committee also sets general policies and reviews the operations of the CRC.
Administration
The CRC presently has a dual administrative locus within MIT. As a research unit, the CRC reports through the Harvard-MIT Division of HST to the vice president for research and associate provost, Professor Alice Gast. However, as a patient-care unit, the CRC is a part of the MIT Medical Department and reports to Dr. William M. Kettyle, the director of the Medical Department. Members of the CRC participate in the Medical Department's activities—e.g., its quality improvement, pharmacy and therapeutics, medical records, and safety committees.
Several years ago the CRC was approached by the General Clinical Research Centers administration of the NIH, which funds this and all other CRCs, and asked to consider becoming a "Network" CRC. This would involve implementing at the MIT CRC some research projects generated at other local CRCs, and, conversely, implementing some of our projects (e.g., those involving very sick patients) at those other centers. Additionally, the CRC would, where possible, coordinate the activities of the core laboratories, nutrition programs, and nursing programs with those of other local institutions, in order to increase their efficiency. The CRC would also use this networking as a platform from which to solicit additional NIH funds, perhaps as a part of a common grant. As a consequence, the CRC has for several years been developing a more structured relationship with the CRC at the Massachusetts General Hospital (MGH), and this expanding relationship has, in fact, been highly successful. To date, thirty MGH protocols have been approved and implemented at the MIT CRC, and three MIT protocols have been implemented at MGH. The senior program staffs at the two institutions continue to meet monthly to anticipate and solve potential problems related to their gradual integration and to streamline the protocol review process; COUHES and its MGH counterpart also work together to evaluate network protocols from the standpoint of safety.
The relationship between the two GCRCs continues to develop and expand. The two centers successfully collaborated on a joint NIH renewal grant application, for five years of support, to start funding in December 2002, when the present MGH NIH grant expires. The score, which the application and site visit received, was the best MIT has received on a five-year renewal. In addition, since the present MIT grant expired November 30, 2001, MGH and MIT jointly submitted an application for one year of funding for the MIT CRC (December 2001 through November 2002) as a dedicated supplement to the MGH grant and the NIH has funded this joint application for the present year. MIT is identified as a "satellite" to the MGH CRC, but will suffer no loss of "sovereignty" or autonomy and, based on discussions with the NIH, no decrease in funding.
Developing this type of "network" relationship with the MGH CRC allows the MIT CRC to solve a chronic problem, i.e., the small and shrinking pool of medical doctors conducting clinical research in this facility, a consequence of the failure, during the last decade, of MIT's academic departments to appoint such people as professors. Most important, it guarantees the longevity of the CRC until such time as the pool again expands, and provides a source of physician scientists to collaborate with MIT biomedical scientists who hold doctoral degrees. The reputations of the two CRCs apparently are excellent, and the strengths of each institution complement those of the other. The CRC also continues to "network" with other Boston-area GCRC's (e.g., BIDMC) and all interested parties agree that the CRC should continue to do so in the future.
Education
The MIT CRC provides formal training in clinical investigation to advanced postdoctoral fellows taking a graduate degree (in clinical research) at Harvard Medical School, and to individual postdoctoral (medical) fellows working with CRC principal investigators and other researchers. These fellows and students utilize the CRC's facilities to initiate research protocols and participate in ongoing projects supervised by senior investigators and faculty. (See section on the Center for Experimental Pharmacology and Therapeutics). The MIT CRC also affords opportunities to MIT undergraduate and graduate students to participate in clinical research projects. In addition, in the spring semester of 2002, Dr. Ravi Thadhani, an assistant program director, taught a formal undergraduate course in clinical investigation. The course was so well received that the decision was made to offer the course again in the spring semester of 2003.
Affirmative Action
The hiring of women and minorities continues to be a high-priority commitment of the CRC. The CRC does have one continuing problem in meeting affirmative action objectives—i.e., attracting qualified minority members. The traditional means of locating such personnel, by advertising and posting positions in local colleges, universities, medical institutions, and minority organizations, have not generated a significant response. Of the seven visiting scientists and scholars appointed by the CRC in AY2002, two were women and one was a minority. The CRC will continue its efforts to increase the pool of qualified minority applicants, as positions become available.
The CRC has been successful in recruiting women and minorities as study subjects. During 2001 approximately 51 percent of all study subjects were women and 11.5 percent of the total study population were black, 5.2 percent Asian, 7.4 percent Hispanic and .4 percent American Indian.
Research Activities
The CRC continues to maintain major commitments to the research activities associated with three clinical areas, each led by a senior professor. These areas are nutrition/metabolism (Vernon R. Young, professor, MIT School of Science), an area in which the CRC constitutes the major locus of MIT's activity, and one that is a traditional component of clinical research centers; neurochemistry/neuropsychopharmacology (Richard J. Wurtman, Cecil H. Green distinguished professor and program director, MIT CRC), which studies the effects of drugs, foods and hormones on brain composition and behavior, the effects of melatonin on sleep, and a set of diseases characterized by affective and appetitive symptoms (i.e., depression, premenstrual syndrome, smoking withdrawal, carbohydrate craving, and obesity), which seem to relate to brain serotonin; and behavioral neuroscience (Emilio Bizzi, Eugene McDermott professor in the brain sciences and human behavior and Lee H. Schwamm, associate professor of neurology at the Harvard Medical School) and neuroendocrinology (Steven K. Grinspoon, assistant professor of medicine at the Harvard Medical School, and Anne Klibanski, professor of medicine at the Harvard Medical School). The behavioral neuroscience component now focuses on strategies for accelerating the return of various brain functions in people who have suffered strokes; the neuroendocrinology component focuses on neuroendocrine concomitants of AIDS, pituitary malfunction, and gender-dependent changes in calcium metabolism.
Groups collaborate on multidisciplinary projects, e.g., obesity, depression, and Alzheimer's disease. The scope of the CRC's activities has expanded broadly. In the past year it also supported research protocols involving, for example, toxicology, pediatrics, psychopharmacology, women's health, HIV, biomedical engineering, and diabetes. Reflecting its evolving interactions with the MGH GCRC, 35 of these projects (out of a total of 67) were directed by investigators whose primary appointments are at the MGH.
During AY2002 the CRC patient census totaled 1,493 outpatient visits and 40 inpatient days. The CRC branch of the NIH had provided, based on prior year's activities, support for up to 2,500 outpatient visits and 84 inpatient days. The decreased census could be explained by the completion of the data-gathering portions of several large projects.
Center For Experimental Pharmacology and Therapeutics
The HST Center for Experimental Pharmacology and Therapeutics (CEPT), based in the MIT CRC, has both an educational and research mission. This center is directed by Dr. Robert Rubin (HST), Osbourne professor of health sciences and technology. Educationally, each year 10 MDs, who have completed their clinical training, enter a two-year program that provides both hands-on research experience and didactic training in clinical investigation and experimental pharmacology. At the end of the two years, after passing a qualifying examination and fulfilling a thesis requirement, the graduates receive a master/medical science degree in clinical investigation from HST. A parallel program for PhD scientists is being established as well. This will involve HST, the Sloan School, the Department of Biology, and the School of Engineering, and will again be centered in the CRC. Research-wise, the emphasis of the CEPT has been in the application of positron emission tomography, magnetic resonance imagery, ultrasound and other measurement technologies to the development of new drugs. With the development of imaging at MIT, these technologies will be greatly facilitated.
Computer Facility
The CRC computer facility provides hardware and software support for the CRC staff andinvestigators and statistical assistance to all researchers. The computer staff continues to develop and upgrade the CRC Operations System with the addition of computer systems for the CRC and investigators. These systems use an ORACLE relational database, which supports the day to day operations of the CRC. The computer staff has also been working with their MGH counterparts to maintain and upgrade the Turbo software package, which has streamlined the protocol application process and NIH annual reporting requirement for both CRCs. In addition, considerable time and effort continues to be spent updating and improving the CRC web site by adding links for MIT IRB protocol applications and expanding the interactive format for the MIT online protocol process. Researchers also continue to make use of the SAS statistical software available on the CRC computer system.
Core Laboratory/Mass Spectrometry Facility
The Core Laboratory specializes in assays that directly support the research efforts of CRC investigators and are not readily available commercially. The most important and complex assays are undertaken by the Mass Spectrometry Facility, where stable isotope tracer analyses are performed. The Mass Spectrometry Facility is a shared instrument facility that allows CRC investigators to conduct human metabolic studies using stable nuclide tracers. Principal areas of investigation concern the regulation of energy substrate metabolism in health and disease, and the regulation of whole body amino acid metabolism, with particular reference to the nutritional requirements for indispensable and conditionally indispensable amino acids. Research at the MIT CRC has made important contributions to the further development of national and international dietary standards and the establishment of sound food and nutrition policies and programs. Studies continue to examine the role of dietary arginine as a precursor of the signal transducer nitric oxide. The novel doubly labeled water (2H218O) method is being used to define the energy requirements for adolescent and elderly subjects, and the factors, which affect these needs. These various investigations offer new basic knowledge about the physiology of human energy substrate and amino acid metabolism and, additionally, make practical contributions to problems in human nutrition.
The Core Laboratory also utilizes high performance liquid chromatography (HPLC) techniques. A Beckman System Gold Amino Acid Analyzer HPLC provides resolution of up to 42 physiologic amino acids. Other HPLC assays include tests for choline, tryptophan, the catecholamines, cytidine and melatonin.
Research Highlights
Dr. Linda Bandini
Dr. Linda Bandini and her colleagues have continued their longitudinal study of the effect of energy expenditure on growth and development in pre-adolescent girls. Annually, subjects visit the CRC for measures of height, weight, and anthropometric measures. In addition, they complete questionnaires regarding their activity and dietary patterns. The study is completed four years after menarche: at study completion the body composition and metabolic rate of the girls are measured in addition to their annual measures. As of June 30, 2002, 154 girls have completed the longitudinal study and only one remains active in the study.
This study will allow the investigators to determine whether reductions in daily energy expenditure or any component of energy expenditure is a risk factor for the development of obesity in adolescent girls. Recently, an article concerning the results of this study entitled "Relationship of Body Composition, Parental Overweight, Pubertal Stage, and Ethnicity to Energy Expenditure Among Premenarcheal Girls" was accepted by the American Journal of Clinical Nutrition.
Dr. Bandini is also investigating the relationship of visceral fat to diet, activity, and hormonal changes in a sub cohort of 40 girls. In this sub cohort, abdominal scans were done at menarche to measure visceral fat and, in these girls, visceral fat is again measured at study completion. These studies will provide information on variables that may influence visceral fat deposition. Determining what factors influence the deposition of visceral fat will provide useful information for the prevention of diabetes and heart disease.
Dr. S. Grinspoon
Dr. Grinspoon and his group have continued to investigate the pathogenesis, clinical phenotype and treatment for HIV lipodystrophy and related metabolic and body composition disorders associated with HIV disease. The HIV lipodystrophy syndrome is a novel metabolic syndrome, characterized by insulin resistance, dyslipidemia and significant changes in fat distribution. Dr. Grinspoon initiated an important collaboration with the Framingham Heart Study to quantify the degree of cardiovascular risk and determine the prevalence of metabolic abnormalities in the HIV lipodystrophy population, demonstrating that almost 45 percent had impaired glucose tolerance, compared with age and BMI-matched patients of the Framingham Offspring study.
Demonstration of significant hyperinsulinemia prompted the first randomized placebo-controlled study of metformin in this population. Results from this study, published in JAMA, showed that metformin treatment could significantly reduce insulin resistance, waist circumference, blood pressure and markers of impaired thrombolysis.
In collaboration with Professor Vernon Young of MIT, Dr. Grinspoon and his group have demonstrated increased lipolysis and a potential role for increased FFA in mediating the insulin resistance. Acute administration of acipimox to such patients doubled insulin sensitivity and decreased FFA. Ongoing MIT GCRC studies, in collaboration with Dr. Colleen Hadigan, are also investigating the utility of dietary manipulation and exercise. Taken together, the studies of Dr. Grinspoon and his group have substantially advanced the understanding of mechanisms causing, and potential treatments, for insulin resistance in HIV lipodystrophy.
Dr. Richard Wurtman
Dr. Richard Wurtman and his colleagues have continued to examine the effects of drugs, foods and hormones on brain composition and behavior. Three sets of pharmacokinetic studies have been performed on compounds, which may subsequently be used to study behavioral or physiological mechanisms. These are: melatonin: an additional study was performed on both 0.3 and 0.6 mg doses, preparatory to a planned multicenter study on the hormone's sleep effects; 5-hydroxytryptophan (5HTP): studies were performed to confirm that it is normally found in the blood, and that administration of low doses without a decarboxylase inhibitor produces dose related increases in plasma 5HTP levels. The compound's effects on stress induced eating will now be studied; uridine monophosphate (UMP) studies are underway to determine whether its oral administration causes dose related increases in plasma uridine levels. If so, studies are planned to see whether it protects against age related memory loss in humans—as it appears to do in rats.
Dr. V. Young
Dr. Young and his colleagues have continued to explore the quantitative aspects of amino acid metabolism in healthy adult humans, with particular reference to their nutritional corollaries. Studies have been completed to the effects of a sulfur amino acid-free diet on whole blood glutathione (GSH) synthesis, showing that GSH production is regulated by the dietary availability of one of its precursors, cysteine. Studies have also been completed on the kinetics and urinary excretion of L-5-oxyoproline, an intermediate of the gamma-glutamyl cycle of GSH synthesis. Both sulfur amino acid-free and glycine-free diets alter the dynamics of oxoproline metabolism and increase the urinary excretion of this intermediate which may, therefore, serve as a potential probe of the status of GSH metabolism in human subjects. Studies have also continued on the kinetic aspects of amino acid metabolism in particular adults. Studies with lysine and threonine as the test amino acids again confirm the hypothesis that the current international requirements values for the indispensable (essential) amino acids in healthy adults are far too low and that the tentative MIT amino acid requirement pattern is an appropriate one for use in practical considerations of adult human protein and amino acid nutrition. These findings and conclusions have major significance with respect to the planning of diets and an evaluation of diets for their amino acid adequacy worldwide. They also have important implications with respect to the planning of agricultural research programs that are directed toward improving the nutritional quality of foods in humans.
More information about the MIT Clinical Research Center can be found on the web at http://web.mit.edu/crc/www/.
Division of Comparative Medicine
The Division of Comparative Medicine (DCM) provides animal husbandry and clinical care for all research animals on the MIT campus. From its inception in 1974, the division has evolved into a comprehensive laboratory animal program that provides a full range of veterinary and surgical support. Additionally, the division has a National Institutes of Health (NIH) grant for training veterinarians for careers in biomedical research. The division also has an active research program funded by numerous R01 grants from NIH. Total personnel in the division now comprises 118 individuals. The division's administrative headquarters along with diagnostic and research laboratories are located on the eighth floor of Building 16. This space is contiguous to the eighth floor of Building 56, which houses quarantine, diagnostic and research space for DCM. The division now encompasses approximately 115,000 square feet devoted to animal research activities. In addition a new vivarium is being planned for the new neuroscience complex.
Facility Management and Animal Care
The average daily census of laboratory animals remained stable during FY2002. Mice remain the primary species used by MIT investigators and represent more than 97 percent of the animal population. The animal facilities support transgenic and gene "knockout" in vivo experiments. DCM now operates a transgenic core and performs a range of transgenic services including in vivo embryo transfer for rederivation of mice with endemic disease which have been imported to MIT from laboratories worldwide, in vitro fertilization, the provision of blastocysts, genotyping of mice and the making of genetically engineered mice. The division has begun to develop expertise in aquaculture and now provides veterinary support for the large zebra fish colonies maintained at MIT. The division received a $660,000 grant from NIH for Improving Institutional Animal Resources. The grant will partially pay for physical improvements to the E25 animal facility as well as the acquisition of additional primate caging and ventilated mouse caging. The animal resource program was recently recertified by the Association for Assessment and Accreditation for Laboratory Animal Care (AAALAC).
Research Activities
Current NIH-funded grants support in vivo study of nitrite carcinogenesis, in vivo study of Helicobacter hepaticus and tumorigenesis, in vivo study of the pathogenesis of inflammatory bowel disease, in vivo study of H. pylori pathogenesis, in vivo study of gastric cancer, in vivo study of heat shock protein and H. pylori pathogenesis and in vivo study of micro-ecology of the gut and the pathogenesis of colitis. Total research funding for the fiscal year was $2.9 million.
FY2002 was the 14th year of the division's NIH postdoctoral training grant that has been funded through year 15. There are currently seven postdoctoral trainees, three of whom are enrolled in the graduate programs in the Division of Biological Engineering. Twenty-six trainees have completed our postdoctoral training program and 23 of them have now passed the board examination of the American College of Laboratory Animal Medicine. Nirah Shomer, while a postdoctoral trainee in DCM, received the Best Research Paper Award for 2001 from the Society for Experimental Biology and Medicine for a paper resulting from her research.
DCM faculty and staff published one book, eight chapters, 25 papers and 28 abstracts in FY2002 and presented numerous research papers at national and international meetings. Dr. Fox is the senior editor of the second edition of Laboratory Animal Medicine, which was published this past winter by Academic Press.
Academic Activities
Dr. James Fox has been appointed to the NIH Scientific Advisory Council of the National Center for Research Resources for 2002–2007. Dr. David Schauer was promoted to associate professor with tenure in the Division of Biological Engineering in 2001. Dr. Mary Patterson, a former postdoctoral trainee, was appointed clinical veterinarian in DCM for large animals on a half-time basis. Both she and Dr. Ihrig passed the board examinations of the American College of Laboratory Animal Medicine this past year. Additionally, Dr. Ihrig is the recipient of a NIH Mentored Clinical Scientist Development Award. She will be involved on a part-time basis as a clinician for DCM's transgenic core. DCM faculty and staff taught two graduate courses in the Division of Biological Engineering (BE.202 and BE.214).
Committee on Animal Care Activities
The web site for the Committee on Animal Care provides required forms, continuing education material and information on the CAC's activities. DCM staff in conjunction with the Committee on Animal Care has developed an online training program. Didactic training sessions for Institute personnel on topics pertaining to the care and use of laboratory animals are also offered. The CAC has also developed an occupational health screen for animal related occupational health issues and periodically sponsors seminars on health issues such as zoonotic diseases. The CAC continued to distribute to other institutions in the United States and abroad two instructional videos, one focusing on the role and responsibilities of Institutional Committees for the Care and Use of Animals and the other focusing on the use of anesthesia in laboratory animals. Both are available to MIT researchers at the division or in the Schering-Plough Library.
More information about the Division of Comparative Medicine can be found on the web at http://web.mit.edu/comp-med/.
Harvard-MIT Division of Health Sciences and Technology
The Harvard-MIT Division of Health Sciences and Technology (HST) brings engineering, science, technology, and medicine to the solution of problems in biology and human health. A successful collaboration that spans more than 30 years between the Massachusetts Institute of Technology (MIT), Harvard University, Harvard Medical School (HMS), area teaching hospitals and research centers, HST is a pioneer in interdisciplinary educational and research programs designed to educate outstanding minds, cultivate leaders, create knowledge, and generate cost-effective preventive, diagnostic, and therapeutic innovations. It is among the largest biomedical engineering and physician/scientist training programs in the United States.
 HST's campus at MIT, the Whitaker College of Health Sciences and Technology (Building E25). Photo by Vikram Kumar. |
Advances in biology and technology are bringing us to an era when diseases can be treated by "engineering" the phenotype of cells and tissues—when cell, tissue, and body functions can be manipulated using strategies that affect genes, cells, and their environment so they behave predictably. Advances in the diagnosis and prevention of disease are inexorably linked to these fundamental changes in our approach to disease management. Unquestionably, success in this area requires professionals with a broad range of skills that spans the domains of science, engineering, and medicine.
HST is dedicated to integrating these disciplines into an educational program that carries engineering and the physical and biological sciences from the laboratory bench to the patient's bedside, and, conversely, bring clinical insights from the bedside to the bench. HST's programs are committed to exploring the fundamental principles underlying disease, to seeking new pharmaceuticals and devices that ameliorate human suffering, and to training the next generation of physicians, scientists, and engineers to do the same. Thus, HST trains physicians with a deep understanding of the underlying quantitative and molecular science of medicine and biomedical research. HST PhD students similarly acquire a deep understanding of engineering and the physical and biological sciences. This unique training is complemented with hands-on experience in the clinic or in industry.
 The Tosteson Medical Education Center in Boston's Longwood medical area. Photo courtesy of Harvard Medical School. |
HST's administrative home is located at the Whitaker College of Health Sciences and Technology at MIT. As one of the five medical societies at Harvard Medical School, HST also maintains an office at the medical school's quadrangle campus in Boston. HST's two co-directors, Martha L. Gray for MIT and Joseph Bonventre for HMS, report to the provost and the vice president for research at MIT, as well as to the HMS executive dean for academic programs and the dean of HMS. Richard N. Mitchell, assistant professor of pathology at Harvard Medical School, serves as the division's associate director and director of student affairs.
Degree Programs
HST currently enrolls approximately 380 students who work with more than 200 faculty and affiliated faculty from the Harvard and MIT communities. Six multidisciplinary graduate degree options are offered, each targeted at students with different backgrounds and goals, each requiring a focused educational and research program, and each offering a different level of clinical training:
- Medical Sciences Program (MD)
- Medical Engineering and Medical Physics Program (MEMP)
- Speech and Hearing Bioscience Technology Doctoral Program (SHBT)
- Radiological Sciences Joint Program (RSJP)
- Medical Informatics Program (MI)
- Clinical Investigator Training Program (CITP)
Research Programs
HST's research programs reflect a mix of cultures in applying the tools of medicine, engineering, and science to problems in human health and medicine. Research initiatives are conducted in three targeted focus areas:
- Biomedical Imaging
- Informatics and Computational Biomedical Sciences
- Regenerative Biomedical Technologies
and two crosscutting research programs:
- Speech and Hearing Bioscience and Technology
- Cardiovascular Sciences and Technology
Highlights
Events
 Richard D. Klausner, MD, delivered the keynote address at HST's 2002 graduation exercises. Photo by L. Barry Hetherington. |
 Eric S. Lander, keynote speaker at the 15th annual HST Forum, talks with Martha L. Bulyk and Laurence R. Young of the HST faculty. Photo by L. Barry Hetherington. |
 Research Day at the Martinos Center for Biomedical Imaging. Photo by Andrew Rolle. |
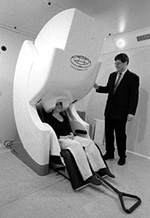 Data from the two new imaging systems at the Martinos Center will produce "movies" of the brain at work. |
 Faculty member Lee Gehrke and MD student Vladmir Vinarsky volunteer at the Greater Boston Food Bank. Photo by Glen Yiu. |
Richard D. Klausner, executive director of the Global Health Program for the Bill and Melinda Gates Foundation, senior fellow of the National Academy of Sciences, liaison to the White House for counter-terrorism, and senior investigator at the National Cancer Institute, delivered the keynote address at HST's graduation on June 5, 2002. Of the 65 graduates who received degrees, 17 graduated with PhD degrees, 33 received the MD degree, and 15 received master's degrees. Eight MD students graduated cum laude. HST's graduating class of 2002 represented 19 states and 13 foreign countries.
The 2002 HST Forum "Beyond the Human Genome" surpassed the number of research posters submitted for previous years (75) and featured keynote speaker Eric S. Lander, professor of biology at MIT and director of the Whitehead Institute/MIT Center for Genome Research. The 15th annual HST Forum welcomed more than 200 attendees on March 14 at the Harvard Club of Boston.
HST's annual John F. and Virginia B. Taplin Awards Symposium was held May 22 to a full audience, including the philanthropists for whom the awards are named. Presentations were given by the 2001 Taplin Fellows: Hugh M. Herr, Martha L. Bulyk, Fiona E. Murray, and Leonid A. Mirny.
The MGH/MIT/HMS Athinoula A. Martinos Center for Biomedical Imaging celebrated its first Research Day at MIT on September 13. "From Structure to Function and Beyond" included a poster session in the Building E25 atrium and provided students and faculty who conduct imaging research an opportunity to meet and interact.
The Martinos Center also dedicated two new state-of-the-art imaging systems at its Charlestown location on April 29. A 7-tesla magnetic resonance imaging system was made possible with the support of the Office of National Drug Control Policy, and a 306-channel magnetoencephalograph system was made possible with the support of the MIND Institute.
Maria S. Judge was appointed HST's new administrative officer and presented her photo exhibition "Toxic, Tattooed and Tougher than Margaret Thatcher: Chronicle of a Year with Cancer" to MIT. The 32nd showing of her exhibit was supported by MIT's Council for the Arts, the Kenneth Schwartz Center, and HST.
HST students, faculty, and staff volunteered to work on the front lines in the community: sixty-five students, staff, and faculty volunteered at the Greater Boston Food Bank on March 6, handling approximately 39,658 pounds of food for redistribution to shelters, soup kitchens, and other providers of meals to the hungry.
Since September, about a dozen HST students, alumni, and staff have spent one Saturday a month at the American Medical Resources Foundation in Brockton, checking and reconditioning medical equipment that will be donated to hospitals and clinics in developing countries.
The HST Visiting Committee convened its biannual meeting in October at MIT and declared HST robust. The meeting provided an opportunity for HMS executive director of academic programs Dennis Kasper, MIT provost Robert Brown, and HMS dean Joseph Martin to demonstrate HST's leading role in transinstitutional education and research programs.
Academics
HST was awarded an NIH National Research Service Award Institutional Training Grant to provide predoctoral training in bioinformatics and functional genomics, thus establishing the new Bioinformatics and Integrative Genomics track (BIG) of the MEMP program. Its mission will be to train quantitative scientists in the biology, engineering, and information science of genomics. The BIG track seeks to train future researchers who must possess the necessary quantitative skills to manage and effectively use the huge amount of data generated by the Human Genome Project. BIG will address bioinformatics in a broad sense—encompassing the full range of work involved to extract functional dependencies from genomic data. Faculty members in the BIG curriculum include nationally recognized leaders from the basic biological sciences, engineering, computer science, genomics, bioinformatics, and epidemiology. Codirectors are Isaac S. Kohane, director of the Children's Hospital Informatics Program and associate professor of pediatrics at HMS, and Gregory Stephanopoulos, professor of chemical engineering at MIT. Both are HST affiliated faculty members.
The medical engineering track of the MIT master of engineering in biomedical engineering (MEBE) was announced. The MEBE program, launched in 2000 with a bioengineering track in the Division of Bioengineering, will be expanded in fall 2002 to include a medical engineering track offered by HST. Through this expanded program, HST and the Division of Bioengineering will prepare students for positions in the medical products, pharmaceutical, and biotechnology industries. The five-year program leads to a bachelor's degree in a science or engineering discipline and a master of engineering in biomedical engineering. The new medical engineering track, which will emphasize engineering applications in systems physiology and cellular, molecular and clinical medicine, will be offered under the auspices of HST beginning September 2002.
HST and MIT's Sloan School of Management jointly established the Biomedical Enterprise Program (BEP), a unique double masters program dedicated to educating a new generation of leaders for tomorrow's biomedical enterprises. The mission of BEP is to equip its graduates to lead the commercialization of new technologies for the innovative diagnosis, prevention, and treatment of human disease in new and established companies. The program will offer a rigorous education in the fundamentals of entrepreneurial businesses, biomedical sciences, and technology. Frank R. Landsberger, co-founder and former chairman of Mojave Therapeutics and founding director of the Office of Science and Technology Development at the Mount Sinai School of Medicine, will serve as executive director of BEP. The curriculum committee was chaired by Fiona Murray, the Michael M. Koerner career development assistant professor of management of technology, innovation and entrepreneurship in the Sloan School.
The Speech and Hearing Sciences Program unveiled a new name: the Speech and Hearing Biosciences and Technology Program. The new name incorporates aspects of technology development that is now an integral part of the program.
New courses were added during AY2002 to enhance HST's curriculum:
- HST 204: Industrial Experience in Medical Engineering and Medical Physics
- HST 398: Introduction to Human Clinical Investigation
- HST 508: Genomics and Computational Biology
- HST 521: Biomaterials and Tissue Engineering in Medical Devices and Artificial Organs
- HST 574: The Engineering of Intermediate Level Sensorimotor Control
- HST 583: Functional Magnetic Resonance Imaging: Data
- HST 918J: Economics of the Health Care Industries
- HST 940J: Bioinformatics—Principles, Methods and Applications
- HST 952: Computing for Biomedical Scientists
- HST 970: Law, Innovation and Entrepreneurship in Health Care
A formal evaluation process for HST PhD courses was established.
The HST Curriculum Committee developed the HST Credo of Professionalism.
In addition to its first HMS DMD student in September 2001, HST admitted 30 MDs, 29 PhDs, three medical informatics master's candidates, and 11 clinical investigator training fellows. Student enrollment hit 385.
During the fall semester, HMS and HSDM launched online access to course materials in MyCourses. John D. Halamka (SM '98), assistant professor of medicine at HMS/BIDMC and associate dean of educational technology, was in charge of developing this system. HST MD student Griffin M. Weber is credited for major input into this password-protected system, which can be found at http://mycourses.med.harvard.edu/.
The MD Admissions Committee received 611 applications; a 17 percent increase over last year. One-quarter (154) of the applicants were interviewed, and 40 of those were offered admission. The gender numbers were equal. The PhD Admissions Committee received 141 applicants. Members of the committee interviewed 41 applicants, offering admission to 25 of them.
Faculty
New faculty appointments:
- Leonid A. Mirny was appointed assistant professor of health sciences and technology in HST. Previously, he was a junior fellow at the Harvard Society of Fellows, where his research involved biophysics and computational biology.
- New HST joint faculty member Martha L. Bulyk was appointed assistant professor of medicine at Harvard Medical School with an appointment in HST. Her research has focused on integrating functional genomics and bioinformatics.
New affiliated faculty members who joined HST in academic year 2001-2002: Lindsey R. Baden; Robert B. Banzett; Stephen C. Blacklow; George M. Church; David Cohen; Clark K. Colton; Randy L. Gollub; David N. Kennedy; Anne Klibanski; Jeffrey D. Macklis; Fiona E. Murray; William C. Quist; David A. Roth; Daniel K. Sodickson; Garrett B. Stanley; Ronald J. Steingard; Clifford J. Tabin; Rav Thadhani; William M. Well.
Faculty promotions:
- Kamran Badizadegan (MD'93), HST affiliated faculty—to assistant professor of pathology at Harvard Medical School/Children's Hospital-Boston
- Jonathan S. Bogan (MD'92), HST affiliated faculty—to assistant professor of medicine at HMS/MGH
- Aziz A. Boxwala, HST affiliated faculty—to assistant professor of radiology at HMS/BWH
- Emery N. Brown, HST faculty—to associate professor of anesthesia at Harvard Medical School/MGH
- Lucila Ohno-Machado, HST faculty—to associate professor of radiology at HMS/BWH
- Steven D. Rauch, HST affiliated faculty—to associate professor of otology and laryngology at Harvard Medical School/Massachusetts Eye & Ear Infirmary
- Daniel Z. Sands, HST affiliated faculty—to assistant professor of medicine at HMS/BIDMC
- After teaching in HST for 29 years, Farish A. Jenkins, Jr., PhD, the Alexander Agassiz professor of zoology at Harvard University and professor of anatomy at HMS/HST, retired as head of HST 010: Functional Anatomy.
- HST co-directors Martha L. Gray and Joseph V. Bonventre wrote the first of three commentaries in the May 2002 issue of Nature Medicine, addressing the recruitment, training, and retention of researchers skilled in translating scientific advances into "something with medical value." Their article, "Training PhD Researchers to Translate Science to Clinical Medicine: Closing the Gap from the Other Side," asserts the effectiveness of this model of training by describing the curriculum of HST's 24-year-old Medical Engineering/Medical Physics (MEMP) Program and its alumni. The authors emphasized the importance of MEMP's four months of clinical training to prepare PhDs for careers in medical research and presented supporting data on MEMP alumni.
- MIT's Clinical Research Center, which is an academic component of HST, and the Massachusetts General Hospital's CRC completed an administrative merger. Each unit continues to operate more-or-less independently with its own director and staff. However, the activities of the two centers are being integrated and a single NIH grant—with a subcontract to MIT—will fund both centers.
- Roger G. Mark, the distinguished professor of health sciences and technology in HST and professor of electrical engineering at MIT, and his wife Dorothy, were named housemasters of the new MIT graduate dormitory at Sidney and Pacific streets.
Milestones
Technical assistant Greg Dancer was one of two recipients of the first Steven Wade Neiterman award. This annual award is given to individuals who possess the following qualities: abilities in collaborative problem solving, in coaching colleagues, and in team building. Dancer was cited for his "motivation to create real solutions and his collaborative and diplomatic spirit."
Database manager/administrative assistant Jennifer Weiss won an MIT 2002 Infinite Mile award for her outstanding achievement, and was recognized for making extraordinary contributions in helping HST carry out its mission.
HST's e-newsletter This Week in HST, or TWiHST, grew to a weekly bulletin with dozens of notices on HST student and community life; information on lectures, workshops, and symposia on HST-related topics; notices of career-related programs at MIT and Harvard; and postings of fellowship and job opportunities. In addition to coverage of HST-specific information, TWiHST has made possible a wide dissemination of broader-based news to the HST community.
In its second year, HST's Irving M. London Society attracted 224 members and raised $184,608, which is a 300 percent increase from FY01 in unrestricted giving. The funds raised through the society provide valuable financial support for HST's students.
HST's Advisory Council convened on November 15 and March 14. The first meeting focused on communications bioscience and technology, and the second looked at the future of bioinformatics at HST.
HST faculty, staff, students, and alumni mourned the passing of:
- Neil S. Ghiso, 31 (MD '01), who died February 11 after a long battle with brain cancer. The Neil Samuel Ghiso Foundation (858 W. Armitage Ave., #111, Chicago, IL 60614) was established in his memory to foster compassionate care for chronically and terminally ill patients and their families through medical education and training.
- Huguette London, wife of founding HST director Irving M. London, who died April 11.
- MIT Professor Emeritus Felix M.H. Villars, a member of the HST faculty since its inception, who died of cancer on April 27.
- Walter A. Rosenblith, 88, MIT provost and Institute Professor emeritus, who died of complications from cancer on May 1. Rosenblith, who served as chair of the MIT Faculty 1967–69, associate provost 1969–71 and provost from 1971–80, was highly influential in the conception, planning and establishment of HST in the late 1960s and early 1970s.
- Lydia A. Cobb, assistant to founding HST director Irving M. London, who died of breast cancer on June 2.
Alumni/ae
Technology Review's June issue featured two HST alumni/ae in its list of 100 brilliant young innovators under 35. Stephen A. Boppart (MD '00, PhD '98), assistant professor of biomedical engineering at the University of Illinois at Urbana-Champaign, was noted for his work on optical-coherence tomography. Jennifer H. Elisseeff (MD '94, PhD '99), assistant professor in the Biomedical Engineering Department at The Johns Hopkins University, was cited for designing a liquid polymer that keeps cartilage cells alive.
Six regional chapter meetings of HST alumni were held across the country:
- Washington, DC/Baltimore: Eighteen alumni/ae from the Washington, DC/Baltimore area met for a reception and dinner on October 10 in Bethesda.
- Los Angeles: The first official HST alumni/ae meeting in southern California, held October 24, was attended by 18 graduates, nearly half of whom work in the Los Angeles area.
- San Diego: A gathering of six alumni/ae was held on October 25.
- Boston: Almost 30 alumni/ae from New England met for a reception and dinner at the University Park Hotel near MIT on November 25.
- New York City: Ten alumni/ae gathered at the Lotus Club in New York City on April 1.
A formal editorial board was appointed for The Connector, HST's alumni newsletter. It will meet twice annually to discuss guidelines and future directions.
Boston Magazine (February 2002) listed five HST alumni/ae among its 170 "Top Docs" in 23 fields:
- Lloyd Klickstein (MD'89) of Brigham & Women's Hospital for autoimmune diseases
- Ralph De la Torre (MD'92) of the Beth Israel Deaconess Medical Center for cardiac surgery
- Michael Fifer (MD'78) of Massachusetts General Hospital for cardiology
- Marc Semigran (MD'83) of Massachusetts General Hospital for cardiology
- Barbara Smith (MD'83) of Massachusetts General Hospital for oncological surgery
Student Honors and Awards
Lea M. Alhilali, second-year MD student, received the 2002 American Society for Artificial Internal Organs (ASAIO) Biomedical Engineering Student Fellowship Award.
Michael R. Folkert received MIT's William L. Stewart Jr. Award, which recognizes outstanding contributions by an individual student or student organization to extracurricular activities and events during the preceding year. Folkert was instrumental in establishing emergency medical training on the MIT campus.
Amy Lee, second-year MD student, received the 2002 HST Student Leadership Award, bestowed annually upon that student who contributes the most to the personal growth and professional development of his or her fellow students in HST.
Eric G. Sheu, first-year MD student, and incoming student Neelaksh K. Varshney were awarded Paul and Daisy Soros Fellowships for New Americans.
Shunmugavelu (Sham) D. Sokka won the Karl Taylor Compton Prize, the highest award presented by MIT to students in recognition of achievements in citizenship and devotion to the welfare of MIT. He was honored for his outstanding contributions to the MIT community as a whole, sustained over a significant number of years.
Howard Hughes Medical Institute award winners included six HST students: Christina L. Boulton, Kevin S. King, Shana E. McCormick, David T. Ting, Vladimir Vinarsky, Nikhil Wagle, and Hao Zhu. Three students received continuing support fellowships: Yvonne Ou, Stephanie N. Misono, and Harris Rose.
Faculty Honors and Awards
David Cohen, HST affiliated faculty and biomagnetism group leader at MIT's Francis Bitter Magnet Laboratory, was appointed associate professor of radiology at HMS and MGH. He is now a member of the Department of Radiology's Nuclear Magnetic Resonance Center for Biomedical Imaging at MGH.
Ernest G. Cravalho, HST founding faculty member and professor of mechanical engineering at MIT, received MIT's Everett Moore Baker Memorial Award for Excellence in Undergraduate Teaching.
Jeffrey S. Flier, HST affiliated faculty and the George C. Reisman professor of medicine at Harvard Medical School and Beth Israel Deaconess Medical Center, was elected to the American Academy of Arts and Sciences and appointed to the newly created position of clinical academic officer and Harvard faculty dean at BIDMC.
Byron J. Good, HST affiliated faculty and professor of medical anthropology in the HMS Department of Social Medicine, was appointed chair of the HMS Department of Social Medicine.
John J. Guinan, Jr., HST affiliated faculty and associate professor of otology and laryngology at Harvard Medical School and the Mass Eye and Ear Infirmary, was corecipient of HST's Irving M. London Teaching Award, which recognizes teaching faculty members who have made outstanding contributions to the training of HST's students.
Roger D. Kamm, HST affiliated faculty and professor of mechanical and bioengineering at MIT, was selected as one of the first eight Cambridge-MIT Fellows for 2001–2002.
Isaac S. Kohane, co-director of HST's track in bioinformatics and integrative genomics and associate professor of pediatrics at HMS and MGH, received the Clifford Barger Excellence in Mentoring Award from Harvard Medical School.
Robert S. Langer, Jr., HST faculty and the Kenneth J. Germeshausen professor of chemical and biomedical engineering at MIT, was awarded the 2002 Charles Stark Draper Prize by the National Academy of Engineering.
M. Charles Liberman, HST affiliated faculty member, director of the Eaton-Peabody Laboratory at the Massachusetts Eye and Ear Infirmary, and professor of otology and laryngology at Harvard Medical School, was awarded HST's Thomas A. McMahon Mentoring Award, which is presented annually to the person who, through the warmth of his/her personality, inspires and nurtures HST students in their scientific and personal growth; and, through honest advice and generosity to all students and colleagues, sets an admirable example of excellence in mentoring.
Steven E. Locke, HST affiliated faculty and associate professor of psychiatry at HMS/BIDMC, was voted president-elect of the American Psychosomatic Society.
Joseph A. Majzoub, HST affiliated faculty and professor of pediatrics at Harvard Medical School and Children's Hospital-Boston, was corecipient of HST's Irving M. London Teaching Award, which recognizes teaching faculty members who have made outstanding contributions to the training of HST's students.
Valerie P. Pronio-Stelluto, HST affiliated faculty and instructor in medicine at HMS and Mt. Auburn Hospital, received the Leo A. Blacklow Teaching Award from Harvard Medical School.
Charles N. Serhan, HST affiliated faculty and professor of anesthesia at Harvard Medical School and Brigham and Women's Hospital, was appointed first distinguished scientist in anesthesiology, perioperative, and pain medicine at Brigham and Women's Hospital.
Myron Spector, HST affiliated faculty and professor of orthopedic surgery (biomaterials) at HMS, received the 2002 Clemson Award for Applied Biomaterials Research, from the Society for Biomaterials.
Jeffrey P. Sutton, HST affiliated faculty and associate professor of psychiatry at HMS/MGH, was appointed director of the National Space Biomedical Research Institute (NSBRI), a consortium of leading biomedical institutions, including HST. Sutton succeeds Laurence R. Young, HST affiliated faculty and the Apollo Program professor of astronautics at MIT, who was honored by the NSBRI for his leadership during its formative years.
Steven E. Weinberger, HST affiliated faculty and professor of medicine at HMS, received the Alpha Omega Alpha Robert J. Glaser Distinguished Teacher Award.
Augustus A. White, III, HST affiliated faculty and professor of orthopedic surgery at Harvard Medical School and Beth Israel Deaconess Medical Center, was appointed master of the Oliver Wendell Holmes Society at Harvard Medical School.
Warren H. Zapol, HST affiliated faculty and the Reginald Jenny professor of anesthesia at HMS, was elected to membership in the Institute of Medicine.
Educational Initiatives
VaNTH Engineering Research Center for Bioengineering Educational Technologies
A consortium of Vanderbilt University, Northwestern University, University of Texas at Austin, and HST, VaNTH is one of 20 Engineering Research Centers (ERC) funded by the National Science Foundation. The ERCs are charged with "providing an integrated environment for academe and industry to focus on next-generation advances in complex engineered systems important for the Nation's future." Unique among the ERCs, the VaNTH ERC focuses on the educational enterprise. Its mission: to "transform bioengineering education to produce adaptive experts by developing, implementing and assessing educational processes, materials and technologies that are readily accessible and widely disseminated. VaNTH will be a working model for how multidisciplinary, multi-institutional groups can define an approach to developing and testing curricula for rapidly evolving knowledge bases." Faculty and researchers in the VaNTH ERC are developing challenged-based learning modules that teach bioengineering content and provide an opportunity for the learner to explore the inter-connections and relationships of the selected concepts. Using the learning sciences philosophy articulated in How People Learn (Bransford, Brown, and Cocking, 2000), each bioengineering module is constructed to be knowledge-centered, learner-centered, assessment-centered, and community-centered. All on equal footing, VaNTH focuses on four research thrusts: the bioengineering domain, learning sciences, learning technology, and evaluation and assessment. Through its educational thrust, VaNTH stresses involvement in research and educational outreach program.
BioMatrix
Founded in the spring semester of 2001 with support from the Alex and Brit d'Arbeloff Fund for Excellence in MIT Education, the BioMatrix mentoring program is designed to create a community of individuals interested in issues related to the life sciences and engineering. This includes undergraduate and graduate students, faculty, clinicians, researchers, and professionals in industry. BioMatrix was headed jointly this year by MIT professor and co-director of HST Martha L. Gray and by Dr. Richard N. Mitchell, associate director of HST at Harvard Medical School. The goal is for this community of like-minded individuals to contribute to the professional and personal growth of students during their MIT experience by providing an opportunity for students to connect with mentors who can help them explore and make career decisions. This year's undergraduate membership was more than 100 students, many of whom became members in their freshman year and continued with the program into their sophomore year. Graduate students include PhD students from several MIT departments as well as MD students in HST. Currently, approximately 50 graduate students are mentors to the undergraduates, and they receive mentoring from practitioner-mentors who include almost 50 basic and clinical researchers, academic physicians, clinicians, health economists and health policy faculty, and biologists in academia and industry. Monthly dinner events have been the backbone of the BioMatrix structure. Each is organized by a student-led subcommittee around a theme selected to appeal to the program's broad membership, but particularly to the undergraduates. This year, BioMatrix members also created one-on-one connections or small group interactions through self-selected activities called BIOs ("BioMatrix Interactions on the Outside"). These offerings, which can be either student- or mentor-initiated, included individual and small-group talks, visits, and professional and social opportunities for BioMatrix members. A third part of the BioMatrix structure involves a web site where members can post profiles, including their academic and personal interests. Members can search profiles to find other members with similar interests. The web site also has an event calendar. The committee work of mentors and students on programming, membership, communications, and assessment has provided a way to engage members in the evolution of BioMatrix and is an excellent opportunity for relationship building. The committee structure gives student members an opportunity to enhance leadership skills. As the program matures, it is expected that additional senior undergraduate members will take on mentoring roles with newer members. BioMatrix provides all undergraduate members the opportunity to build close relationships with faculty and other professionals outside the academic context and to work with mentors in professional settings. As it grows, BioMatrix will seek to establish MIT's prominence in attracting students with career goals in the life science arena.
Research Achievements
The research of the HST core faculty and research staff covers a wide spectrum of biomedical areas. In addition to laboratories at HST, MIT, Harvard University, and the Harvard Medical School, research collaborations include several HMS teaching hospitals in the Boston area (including Massachusetts General Hospital, Brigham and Women's Hospital, Beth Israel Deaconess Medical Center, Dana Farber Cancer Institute, and Children's Hospital).
Regenerative Biomedical Technologies
The objective of HST's efforts in regenerative biomedical technologies is the cost-effective replacement of cell, tissue, and organ function. Toward this end, HST researchers apply the rigors of physical sciences to problems in medicine and biology. In particular, this work seeks to understand, harness, and engineer tissues, cells, and molecules. A principal site of this effort is the Harvard-MIT Biomedical Engineering Center, located on the MIT campus. There, seven resident faculty members supervise a scientific staff of 90. An additional 70 affiliated faculty at MIT and HMS use the center's extensive facilities and resources, which support work in a broad range of integrated sciences from molecular and cell biology to animal physiology.
Elazer R. Edelman (HST '83) is the Thomas D. and Virginia W. Cabot associate professor of HST and director of the Harvard-MIT Biomedical Engineering Center. Dr. Edelman and colleagues in his laboratory use elements of continuum mechanics, digital signal processing, molecular biology, and polymeric controlled release technology to examine the cellular and molecular mechanisms that transform stable coronary artery disease to unstable coronary syndromes. Tissue-generated cells, for example, deliver growth factors and growth inhibitors for the study and potential treatment of accelerated arterial disease following angioplasty and bypass surgery. The laboratory holds patents for drug delivery devices, tissue engineered implants, and new drug formulations.
Michael S. Feld heads the MIT Laser Biomedical Research Center, an NIH Biotechnology Resource Center housed in the MIT Spectroscopy Laboratory, which develops basic scientific understanding, new techniques, and technology for advanced biomedical applications of lasers. Fluorescence, reflectance, near-IR Raman, light-scattering spectroscopy, and low coherence interferometry are being used for histological and biochemical analysis of tissues, diagnosis and imaging of disease, and cell biology applications. Clinical studies are conducted with researchers from the Cleveland Clinic Foundation, Medical University of South Carolina, Brigham and Women's Hospital, Metrowest Hospital, Beth Israel Hospital and New England Medical Center. Clinical studies using trimodal spectroscopy, the combined application of intrinsic fluorescence, diffuse reflectance, and light scattering spectroscopies, have demonstrated successful diagnosis of dysplasia in Barrett's esophagus, the urinary bladder, adenomatous polyps, the oral cavity and the uterine cervix. Light scattering spectroscopy was used to measure and image subcellular structures much smaller than an optical wavelength. Novel low coherence interferometry techniques, which use two harmonically related wavelengths to measure optical phase, have been developed. Exceedingly small refractive index and length changes, tomographically mapped, were used to study structure and dynamics of cellular organelles. Raman spectroscopy was used to measure blood analytes with clinical accuracy and identify morphology of breast lesions. The experimental and theoretical work of this program is advancing new laser diagnostic technologies in the fields of medicine and cell biology.
Hugh M. Herr and colleagues at MIT's Leg Laboratory seek to understand how the mechanics, energetics, and control of locomotion are determined by speed, animal size, and fundamental forces such as gravity and inertia. Towards this goal, they are testing hypotheses that integrate the mechanics, energetics, and control of locomotion, and have developed morphologically realistic, physics-based computer models that predict important features of mammalian trotting and galloping. Their virtual models span nearly three orders of magnitude in body size: two horses, a goat, two dogs, and a chipmunk. Using these virtual creatures, they are testing the effects of systematic changes in structure and control parameters. In another project, an auto-adaptive knee prosthesis is being developed for trans-femoral amputees that will move naturally at all locomotory speeds and perform equally well for all amputees. Using state of the art prosthetic knee technology, a prosthetist must pre-program knee damping values until a knee is comfortable and safe to use. Their knee prosthesis automatically adapts to the amputee without pre-programmed information of any kind from either amputee or prosthetist. The adaptation scheme successfully controls early stance resistance, swing phase peak flexion angle and extension damping, suggesting that local sensing and computation are all that is required for an amputee to walk in a safe, comfortable and smooth manner. Finally, in work with the Biomechatronics Group, Dr. Herr is developing small robots actuated by animal derived muscle tissue. In this investigation, muscle tissues are specifically engineered for machine actuation. Genetic, chemical, electromechanical, and temperature interventions are used to enhance muscle robustness and contractile function in vitro. Two types of muscle tissue are being examined: native and cultured tissues from genetically modified mice and native whole muscle from non-mammalian sources such as marine invertebrates. Once engineered, the contractility and robustness of these tissues will be characterized and comparisons will be made to current artificial muscle technologies.
Robert S. Langer, Jr., a pioneer in biomedical and chemical engineering, is studying new ways to deliver drugs, including a new microchip that can deliver drugs in a pulsable fashion. He is also researching tissue engineering and has created new approaches for creating blood vessels, cartilage, and many other tissues. He has also developed biomaterials for medicine, including plastic that slowly dissolves and releases therapeutic drugs directly to tumors. In 1996, this led to the first new treatment for brain cancer approved by the FDA in more than twenty years.
Roger G. Mark, distinguished professor of health sciences and technology, together with colleagues at Beth Israel-Deaconess Medical Center, Boston University, and McGill University, continue to develop the new NIH-funded "Research Resource for Complex Physiologic Signals." The resource investigates cutting-edge physiologic signal processing techniques, and freely distributes extensive archives of annotated physiologic data and signal processing software to the international research community via the Internet (http://www.physionet.org/). Dr. Mark's group also is developing computational cardiovascular models to better understand orthostatic intolerance induced by space flight and is exploring innovative approaches to "intelligent" patient monitoring.
Frederick J. Schoen has made major investigative contributions to understanding the problems of currently available prosthetic devices and patient-management strategies. He has identified, elucidated the mechanisms of, and solved several of the critical problems associated with the biomaterials and devices used clinically, especially substitute heart valves. His approaches have used basic biology, evaluations of clinical implants that have failed, and industrial development studies of new and modified configurations and biomaterials. Ongoing investigations are focusing on cell-extracellular matrix interactions in the mechanisms of native heart valve degeneration and as determinants of the structure-function-quality correlations in heart valves fabricated by tissue engineering methods.
Martin L. Yarmush and colleagues are contributing to several fields, including tissue engineering, gene therapy and nucleic acid biotechnology, genomic and proteomic technology, and metabolic engineering. Drs. Yarmush, Mehmet Toner, and Ronald G. Tompkins are collaborating on one of the world's leading programs to establish a liver support device using hepatocytes and microfabrication techniques. In addition, in the area of tissue engineering, Drs. Jeffrey R. Morgan and Yarmush are developing the next generation of skin substitutes using genetically modified cells. In the areas of gene therapy and nucleic acid biotechnology, Dr. Yarmush's laboratory, together with those of Drs. Morgan and Arul Jayaraman, are investigating rate limiting aspects of gene therapy and antisense therapy. Drs. Yarmush, Toner, Morgan, and Jayaraman are also collaborating on a new platform for monitoring real time gene expression using a living cell microarray, which can provide minute by minute information in a massively parallel format. Finally, Drs. Yarmush and François Berthiaume are using the tools of metabolic engineering to investigate the complex metabolic changes that occur in chronic disease and major injury.
James C. Weaver, HST senior research scientist, and his research group, have extended their development of a new computer simulation method to image-based modes at the cellular level, such that realistic models of human skin stratum corneum and other tissues can be created and solved for "in silico" assessment transport of beneficial and hazardous chemicals.
Lisa E. Freed, HST principal research scientist, supervises a research team working on tissue engineering. Her research interests include cell and developmental biology, biomaterials, and biomedical engineering, and in particular the integrated use of cells, three-dimensional scaffolds, and bioreactors to engineer functional skeletal and cardiovascular tissues. The goals are to improve basic understanding of tissue development through controlled in vitro studies and to generate clinically useful tissue equivalents. She also participated in teaching Biomaterials and Tissue Engineering in Medical Devices and Artificial Organs (HST 521).
Gordana Vunjak-Novakovic, HST principal research scientist, is supervising research teams working on tissue engineering of skeletal and cardiac tissues, and biological research in space. Her research interests include tissue engineering, bioreactors, and transport phenomena in living systems, and in particular the integrated use of cells, biomaterials and bioreactors in quantitative studies of cell function and tissue development. She is serving as the science lead of the design and testing of the cell culture system for the International Space Station. Her teaching responsibilities include Biomaterials and Tissue Engineering for Medical Devices (HST 588, course codirector), Quantitative Physiology of Cells and Tissues (6.021J, recitation instructor), and Biomaterials and Tissue Engineering (ChE 164 at Tufts University, lecturer).
An important scientific challenge in regenerative biomedical technologies research is to understand biological complexity: how life and cellular function emerge from the interactions of these different components. HST's work in this area aims to develop entirely new analytical tools and computational models needed to describe the nonlinear emergent behavior of complex biological systems.
Joseph V. Bonventre (HST '76), HST codirector, studies the mechanisms of cellular and tissue injury and repair, particularly as related to ischemic injury to the kidney. Recent studies have focused on the role of inflammation and adhesion in the pathophysiology of acute renal failure. A novel adhesion molecule, KIM-1, has been cloned that is expressed at very high levels during the recovery phase of acute renal failure and in models of chronic renal disease as well as in a number of human kidney diseases including polycystic kidney disease. This molecule is shed from the cell membrane, and it appears in the urine of patients with kidney injury at an early stage of the disease process. Using PCR-based subtraction techniques and bioinformatics, many additional genes whose regulation is altered during repair have been identified. Many of these represent potential targets for therapeutic interventions to prevent or treat kidney injury. Bone marrow derived stem cells have differentiated to epithelial cells replacing injured kidney cells. Approaches are being explored to facilitate this process and potentially regenerate the tubular epithelium and restore function to a failing kidney. Transcription factors are important determinants of the cellular repair processes after an ischemic insult to the kidney. A novel kidney-specific zinc finger transcriptional repressor, Kid-1, whose expression is regulated in renal ontogeny and by ischemia/reperfusion was cloned and characterized. The Kruppel Associated Box-A (KRAB-A) motif of this and other zinc finger proteins was identified as a common repressor motif. A transcriptional repressor, KRIP-1, that interacts with KRAB-A has been cloned. A new family of proteins that associates with KRIP-1 (Trip-Br family) have been characterized which interact with E2F/DP1, two critical proteins for cell cycle regulation. A second major focus of the lab is phospholipase A2 (PLA2) and the role of this family of enzymes on acute tissue injury, apoptosis, signal transduction and nuclear events including transcription. Using the yeast two-hybrid system, a nuclear protein that interacts with the cytosolic 85 kDa cPLA2 has been identified. A cPLA2 knock-out mouse has been created to study the function of PLA2s in signal transduction and renal, respiratory, gastrointestinal and neurological disease. Gene therapy approaches with adenovirus are being used.
George Q. Daley (HST '91) is investigating the signaling pathways that allow the BCR/ABL oncoprotein to induce leukemia. His laboratory has demonstrated that a novel class of pharmaceutical agents called farnesyl transferase inhibitors have potent activity against BCR/ABL-induced leukemia and active clinical testing of these agents are underway. His lab has also demonstrated that hematopoietic stem cells develop from pluripotent embryonic stem (ES) cells that are differentiated in culture, and is the first to combine nuclear transfer cloning and ES cell differentiation (therapeutic cloning) to treat a genetic disease in the mouse, an important step towards using ES cells for cellular therapies.
Lee Gehrke, professor of health sciences and technology, studies the replication and assembly of viruses that use RNA as their genetic material. Important biochemical processes that allow viruses to replicate depend on docking interactions between RNA and protein molecules. Dr. Gehrke's laboratory is focused on identifying these docking signals, an effort that will facilitate therapeutic approaches for blocking virus replication and assembly. The research has led to the molecular identification of amino acids and nucleotide sequences that are crucial for forming the RNA-protein interactions; moreover, the work also suggests the shape or conformation of the molecules changes upon binding. Another aspect of his work is learning how viruses are able to gain an advantage over the infected host cell in expressing their own genetic information. Nucleotide signals in a viral messenger RNA have been identified that give the virus a competitive advantage, and the lab is now working to elucidate the detailed mechanism.
Richard N. Mitchell, HST's associate director, researches the mechanisms underlying acute and chronic rejection in solid organ allografts, with specific emphasis on heart transplants. His work runs the gamut from mouse transplant models to human clinical transplantation, and is focused on understanding the specific immunologic pathways that drive rejection and ultimately graft failure. His lab is particularly interested in the mechanisms that induce the process of "chronic rejection" whereby the vessels in transplanted hearts become progressively more occluded until the grafts get starved for blood and die. The research may have much broader applicability, since the inflammatory mediators that drive the occlusive process in transplanted hearts may also be involved in mediating the vascular wall thickening that characterizes more "typical" atherosclerosis. Dr. Mitchell's laboratory uses several genetically engineered mice (so-called "knock-out" mice), which are either deficient in cell surface molecules that promote the cellular cross talk necessary to promote rejection, or which lack particular "cytokine" mediators or their receptors. In collaboration with other members of the HST community, such as Drs. Elazer Edelman and Andrew Lichtman, Dr. Mitchell has been evaluating new interventions to prevent the chronic vascular pathology. His group has also developed collaborations with several pharmaceutical firms such as Schering-Plough, Bristol Myers-Squibb, and Novartis.
Jane-Jane Chen, principal research scientist in HST, studies the regulation of hemoglobin synthesis and erythropoiesis by the heme-regulated eIF-2 alpha kinase (HRI). Her group has knocked out the HRI gene in mice, and established that HRI, which is expressed predominantly in erythroid cells, regulates the synthesis of both a and b globins in red blood cell precursors by inhibiting the general translation initiation factor eIF2. This inhibition occurs not only when the intracellular concentration of heme declines, but also occurs when cells are under various cytoplasmic stresses. HRI is responsible for the adaptation to well tolerated microcytic, hypochromic anemia occurred in iron deficiency. Our recent studies of mice with compounded HRI and b-globin or ferrochelatase deficiencies demonstrate that HRI can be a modifier gene and affect the severities of b-thalassemia and erythropoietic protoporphyria in mice. These data have significance for further understanding the physiological role of HRI as a guardian of hemoglobin synthesis and red blood cell production against various stresses.
HST's research in regenerative biomedical technologies also encompasses investigations into therapeutics and clinical human studies, which provide important insight into disease mechanisms and new diagnostic procedures. The opportunities available through MIT's Clinical Research Center, the Harvard Medical School teaching hospitals, and HST's Clinical Investigator Training Program significantly enhance the division's research and educational initiatives and further enable translational efforts from bench to bedside.
Robert H. Rubin, the Gordon and Marjorie Osborne professor of HST, has spent much of his clinical career studying and caring for transplant patients. Among his accomplishments are the development of new strategies for preventing the most important infections, particularly those due to viruses and fungi; the establishment of the link between certain viral infections and allograft injury and the development of certain malignancies; and the development of novel antimicrobial approaches that are effective not only in transplant patients, but also in such other immunocompromised patient populations as those with AIDS and cancer. In 1999, he was named to the chairmanship of the Infectious Disease Section of the International Transplantation Society and assumed the position of editor in chief of the journal, Transplant Infectious Disease. More recently, he has taken on the responsibility of developing Internet-based educational programs for international use and in 2002 worked as editor-in-chief for Good Clinical Practices in Clinical Research and Fungal Infections: Grand Rounds. As director of HST's Center for Experimental Pharmacology and Therapeutics, Dr. Rubin has pioneered the application of positron emission tomography, magnetic resonance imaging and spectroscopy, and other measurement technologies to the development of new drugs, including those designed for the transplant patient. In addition to his other responsibilities, Dr. Rubin is associate director of the Division of Infectious Diseases at the Brigham and Women's Hospital, charged with the responsibility of directing the clinical service and the clinical research program.
Drs. Rubin and Alan C. Moses head the two year Clinical Investigator Training Program, a joint effort of the Beth Israel-Deaconess Hospital, HST, and Pfizer, Inc. Trainees gain direct experience in clinical investigation and a strong foundation in the statistical and computational sciences, biomedical ethics, principles of clinical pharmacology, in vitro and in vivo measurement techniques, and aspects of the drug development process. After fulfillment of thesis requirements and successful performance on a qualifying exam, graduating trainees receive a MMSc degree in clinical investigation from HST.
Richard J. Wurtman, program director of MIT's Clinical Research Center, also conducts research into Alzheimer's disease. A generally held—if unproved—view of Alzheimer's is that dementia results from toxic effects of an abnormal protein, called amyloid, which is a polymer of small fragment (A-beta) of a protein (APP) produced normally in all cells. Hence, a major goal of researchers working to treat this disease is to find drugs that will decrease the formation of A-beta from APP and increase the production of APPs other major metabolite APPs ("soluble APP"). Dr. Wurtman's laboratory has shown that both the synthesis of the APP and the proportions of it that are broken down to A-beta or soluble APP are under the control of the particular neurotransmitters and "second messengers" they generate. Thus, by using drugs that act on these receptors, it should be possible to block the formation of APP and all its metabolites, or promote the formation of soluble APP and suppress A-beta. This has been demonstrated in tissue culture and is in the process of being demonstrated in animal models of Alzheimer's. The next step, probably involving industry collaboration, involves devising a treatment to decrease the amount of amyloid in the Alzheimer's disease brain. This treatment may conceivably ameliorate the dementia of the disease.
Biomedical Imaging
Biomedical imaging is a relatively young field that enables physicians and scientists to "see" and better understand tissue and organ function. It provides investigators with the ability to visualize the structure of tissues and to capture their function on film. One type of biomedical imaging is magnetic resonance imaging (MRI), also known as nuclear magnetic resonance (NMR). Using electromagnetic fields and radio waves to read minute shifts in the magnetic alignment of protons in soft tissue such as the brain, it involves the collaboration of engineers, computer scientists, neuroscientists, and physicians. An important advance called functional MRI (fMRI) shows how living tissues are functioning in real time. For example, fMRI can make a 100-millisecond scan every few seconds to detect variations in regional blood flow within the brain to signal sight, hearing, thinking, or feeling. Combining many fMRI scans makes a real time "movie" of functioning organs, a technique that has been especially useful in cognitive neuroscience and psychology.
Emery N. Brown devotes his research to statistical modeling of problems in neuroscience. Working jointly with colleagues in MIT's Brain and Cognitive Sciences Department, he is developing statistical signal processing techniques to analyze how neural systems encode information about relevant biological stimuli in their ensemble firing patterns. The techniques involve signal processing methods based on point process filtering, and have been successfully applied to the study of how ensembles of pyramidal cells in the rat hippocampus encode the animal's representation of its spatial environment. In collaboration with members of the MGH NMR Center, Dr. Brown is developing statistical methods to characterize the dynamic properties of functional magnetic resonance imaging (fMRI) signals. A forthcoming application of these methods will be to study anesthesia induced loss of consciousness monitored with fMRI. In collaboration with colleagues at the Brigham and Women's Hospital, Dr. Brown has developed statistical models to measure precisely the period of the human biological clock and to characterize the properties of circadian and neuroendocrine rhythms.
Martha L. Gray (HST '86), HST co-director, and collaborator Deborah Burstein (HST'86) use magnetic resonance to measure the composition and functional integrity of cartilage. Over the last century, little progress has been made in developing effective preventative and therapeutic strategies for arthritis, other than total joint replacement. Among the challenges limiting progress has been the fact that there were no nondestructive means to visualize cartilage. Pioneering work by this team has yielded a clinically feasible method that is now employed in pilot studies. Work with Genzyme's Carticel™ product revealed an apparent improvement by 18 months after surgery. An evaluation of dysplastic hips revealed composition correlates with pain. These are examples of information that were previously unavailable. Clinicians and researchers have had to struggle to understand and treat diseases they could not see until significant cartilage destruction had occurred. This situation has the potential to improve dramatically with the method Drs. Gray and Burstein have pioneered. In the past year, this method has been used to demonstrate nondestructively biochemical alterations in intact human cartilage. In addition, pilot clinical studies have begun to demonstrate subtle alterations in cartilage, which may be amenable to early pharmacologic intervention
Bruce R. Rosen (HST '84) is director of the Nuclear Magnetic Resonance Center at the Massachusetts General Hospital and the MGH/MIT/HMS Athinoula A. Martinos Center for Biomedical Imaging. The Martinos Center fosters biomedical imaging research that spans scientific disciplines from basic research to clinical investigation and develops medical applications for these new technologies. This new biomedical imaging center is a partnership between HST, Massachusetts General Hospital, and the Harvard Medical School. Its mission is to build the next generation of functional imaging tools; to apply these tools to biologically, neurologically, and clinically relevant problems; to train physical, biological, and clinical scientists; and to provide a hub for interdisciplinary collaborations between Harvard, MIT, and other institutions worldwide. Dr. Rosen is well known for his contributions in the area of "functional" imaging—that is, magnetic resonance images of the brain in which areas having some functional activity (e.g., visual cortex) are highlighted by receiving increased blood flow. The techniques he and his colleagues have developed are being used by hospitals throughout the world to evaluate patients with stroke, brain tumors, dementia, and other mental illness. Recent work has focused on the fusion of functional MRI data with information from other modalities, including very high temporal resolution signals using magnetoencephalography (MEG) and noninvasive optical imaging.
Informatics and Computational Biomedical Sciences
Knowledge discovery and its dissemination in health care have been deeply influenced by recent advances in computer science and engineering. Medical and biological informatics is the use of computer technology to extract, transport, and manage information from medical and biological data, and to model and support human decision making in clinical and biological domains. Research challenges include: deducing and mapping genomic structure, predicting structure and function of proteins, representing medical knowledge for modeling diagnostic and prognostic decision making processes, extracting new information from large clinical and biological data sets, building comprehensive electronic medical records (EMR) and clinical information systems, interfacing monitoring devices and the EMR, assuring privacy and confidentiality in medical transactions, analyzing and manipulating images, recognizing patterns of disease progression, analyzing costs and benefits related to medical use of information technology, computer aided instruction, and utilizing the Internet for providing education and health care services.
Martha L. Bulyk uses computational and experimental approaches to study transcriptional regulatory networks in model organisms and in the human genome. She and her colleagues use various computational methods to identify likely DNA regulatory elements in the human genome. They are also applying and developing new microarray technologies for the high-throughput characterization of the binding specificities and regulatory roles of transcription factors (TFs). Predicted regulatory elements and corresponding TFs will be verified using these technologies. These studies will permit a better understanding of the locations and organization of regulatory DNA elements in higher eukaryotic genomes, and will aid in understanding the regulatory complexity resulting from combinatorial interactions of TFs. These data will also permit the development of more accurate algorithms to predict DNA regulatory elements in the human genome. Furthermore, the combination of these data with mRNA expression analysis, protein interaction databases and prior genetic and biochemical data in the literature will allow the construction of more detailed connectivity maps of transcriptional regulatory networks. Dr. Bulyk's laboratory will make its data publicly available, enabling other researchers to focus their efforts on those genomic regions most likely to contain regulatory elements.
Robert A. Greenes, director of HST's Biomedical Informatics Training Program, established the Decision Systems Group (DSG) at Brigham and Women's Hospital in 1978 to pursue the application of information technology in health care education and decision making. Dr. Greenes has a 38 year history of work in the area of biomedical informatics. The DSG lab, which includes physicians, computer scientists, database experts, and specialists in graphics and educational technology, has a major focus on developing means to enhance decision support and education and integrate these capabilities into clinical practice. Primary technologies involve knowledge representation, machine learning, decision science, and information retrieval. The DSG is also working in the area of bioinformatics, particularly in the development and application of machine learning, classification, and prediction methods for the analysis of DNA microarray data and of flow cytometry data characterizing cellular proteins. The HST-based Biomedical Informatics Training Program, which Dr. Greenes directs, is funded by an institutional training grant from the National Library of Medicine. This year, the program received a competitive renewal award with an annual budget of approximately $2 million.
Isaac S. Kohane, director of HST's bioinformatics and integrative genomics track, is involved in leading multiple collaborators in bioinformatics and the functional genomics of tumorigenesis, neurodevelopment, neuroendocrinology, and transplantation biology. He also leads several national efforts in distributed clinical computing, biosurveillance, and decision support with an emphasis on the use of cryptographic methods to protect patient privacy.
Leonid A. Mirny, assistant professor of health sciences and technology, conducts research in computational structural and system biology. His efforts in computational structural biology are focused on development of novel computational tools to analyze and predict structures of proteins, their complexes and protein-DNA interactions. His system biology projects integrate large scale analysis of proteomic and genomic data with molecular simulations of proteins. He is interested in developing stochastic computational models of genetic and biochemical networks, cellular regulation and signal transduction. His research is aimed at efficient extraction of biological knowledge from experimental genomic and proteomic data as well as at fundamental understanding of molecular mechanisms involved in cellular regulation.
Lucila Ohno-Machado investigates machine-learning techniques to extract information from databases, especially in the form of predictive models for prognosis. She has used special methods to predict survival for patients with certain conditions, to assess the risk of myocardial infarction in certain populations, to predict ambulation for patients with specific kinds of spinal cord injuries, and to predict outcomes for invasive interventions. Her research is focused on the development and evaluation of models involving binary outcomes using mixtures of machine learning and statistical models. These models can be based on clinical and genomic data. She is interested in deploying practical models for direct use by patients, physicians, and health care managers, so that they can make decisions that are more informed. An example in this area is a project that uses artificial intelligence techniques for dealing with uncertainty to select suitable clinical trials for patients with certain types of breast cancer. Other areas of interest include decision support to help patients recognize early symptoms of a heart attack, remote monitoring, and other uses of information technology to facilitate collection and analysis of biomedical data.
Speech and Hearing Bioscience and Technology
In a world with increasingly complex societal interaction, there is an imperative need to understand how humans communicate, and what can be done to assist those with impaired abilities to produce speech or to perceive auditory signals. HST's Speech and Hearing Sciences research focuses on the biological and physical mechanisms underlying human communication by spoken language. The processes addressed by these sciences include the physical acoustics of sound and the perceptual and neurophysiological bases of hearing, as well as the cognitive and linguistic levels of processing by talkers and listeners. This research, and its application to human needs, is inherently an interdisciplinary activity.
Understanding the process of receiving and translating speech sounds has been the focus of several HST researchers, including Dennis M. Freeman. Via a bundle of microscopic hairs, sensory cells in the inner-ear sense sound-induced motions of inner-ear structures, which trigger neural messages that relay information about sound to the brain. Dr. Freeman and his colleagues have measured the motions of a lizard's inner-ear sensory cells in response to sound stimulation. They have also measured mechanical properties of the tectorial membrane, an essential tissue that mechanically stimulates hair cells. In this work, they have obtained the first quantitative measurements of the bulk modulus, fixed charge concentration, and point impedance of the tectorial membrane. In their related microelectromechanical systems (MEMS) work, his group developed a Mirau interferometric imaging system to characterize MEMS motions and used it to measure dynamic profiles of flexible MEMS (optical gratings). This dynamic profilometer for MEMS is likely to have important applications in many MEMS labs. In the area of synthetic aperture optics, in addition to building a 50-beam synthetic aperture light projector using 488 nm light, his group also built a surface-acoustic-wave (SAW) light modulator and used it to break a single incident beam at 193 nm into hundreds of output beams with controlled amplitudes and phases. The SAW optical modulator is likely to have wide application as a general light modulator.
M. Charles Liberman, director of the Eaton-Peabody Laboratory at the Massachusetts Eye and Ear Infirmary, studies the neurobiology of hearing and hearing loss in animals and humans. His work on normal hearing investigates the structure and function of the four major classes of afferent and efferent neurons connecting the inner ear with the brain. Notable recent progress includes the discoveries that sound-evoked activation of one of the efferent feedback pathways protects the inner ear from permanent acoustic injury and that inter-subject variation in the strength of this feedback reflex can predict susceptibility to hearing damage in noisy environments. His work on hearing loss is currently focusing on prevention of presbycusis, or age-related hearing loss. His laboratory has recently demonstrated a genetic manipulation in mice, which dramatically minimizes age-related hearing loss as well as the age-related loss of sensory cells. The molecular mechanisms underlying this protective effect are now under investigation.
John J. Rosowski is codirector of the Wallace Middle-Ear Research Unit of the Eaton-Peabody Laboratory at the Massachusetts Eye and Ear Infirmary. He and his colleagues strive to understand how the structures of the external and middle ear affect what we hear by measuring the function and structure in normal and pathological ears. On the order of one percent of the human population suffers from some form of chronic middle-ear disease, and thousands of surgeries are performed annually at the Massachusetts Eye and Ear Infirmary alone to control these diseases and restore hearing. The Wallace unit is investigating the clinical utility of laser-Doppler measurements of sound-induced middle-ear velocity in patients and human subjects in order to diagnose middle-ear disease and evaluate post-surgical results. Preliminary results indicate certain hard to diagnose ossicular abnormalities can be differentiated by vibrometry, and that the reasons for a large percentage of middle-ear surgical failures can be determined by this procedure.
Bertrand Delgutte and his colleagues seek to understand the neural mechanisms for perception of sounds. A focus of their research in the past year has been to improve processors for cochlear implants, prosthetic devices that electrically stimulate the auditory nerve in order to restore hearing in the profoundly deaf. Most current processors only deliver information about the slowly varying temporal envelope of sounds, and discard the rapidly varying, fine time structure. To assess the relative perceptual importance of these two components of sounds, HST graduate student Zachary Smith, psychophysicist Dr. Andrew Oxenham and Dr. Delgutte created novel chimeric sounds combining the temporal envelope of one sound with the fine structure of another. They found that speech reception depends primarily on the envelope, while melody recognition and sound localization depend on the fine structure. This finding suggests that modifying cochlear implant processors to deliver fine structure information would improve music appreciation and, for patients implanted in both ears, speech reception in noise. In related work with HST graduate student Leonid Litvak and Dr. Donald Eddington, he showed that a new strategy for delivering fine structure information in cochlear implants gives a more natural representation of sounds in the auditory nerve than do existing processors. The effectiveness of this strategy is now being tested in human patients.
Cardiovascular Science and Technology
HST's cardiovascular research is dedicated to developing innovative diagnostic and therapeutic technologies and medications that will have an impact on clinical care within a ten-year period. Traditionally, academic institutions have focused on long-term basic research, whereas the research efforts of pharmaceutical companies, as well device and equipment manufacturers, have focused on development. HST's cardiovascular investigations focus on novel ideas that bridge this gap between basic research and applied development. In order to achieve translational research and to train new investigators in this field, HST builds on its strong ties to academic and clinical laboratories, including the Boston Heart Foundation, and develops new ties with industry. In cooperation with these partners, HST's cardiopulmonary effort will be able to provide products designed for a patient's specific phenotype when developing new technologies and drugs.
The laboratory of Richard J. Cohen (HST '76), Whitaker professor of biomedical engineering, is involved in the development of cardiovascular diagnostic and therapeutic technologies. One of the technologies developed in his laboratory is the noninvasive measurement of microvolt level fluctuations in electrocardiographic signals to identify individuals at risk for sudden cardiac death from heart rhythm disturbances. This technology, called the measurement of microvolt T-wave alternans, has been commercialized by a company that Dr. Cohen helped found—Cambridge Heart, Inc. The technology has been successfully tested in a wide range of international clinical trials and is currently being introduced into widespread clinical practice. Under sponsorship of the National Space Biomedical Research Institute, Dr. Cohen is using this technology to determine whether long-duration space flight increases the risk of life-threatening heart rhythm disturbances. Dr. Cohen's laboratory was also involved in demonstrating the effectiveness of a new pharmacologic countermeasure to another adverse effect of space flight on the cardiovascular system. After long-duration space flight, astronauts develop orthostatic hypotension, meaning that their blood pressure drops when they attempt to sit or stand. This effect may be so severe as to cause syncope or fainting. Dr. Cohen's laboratory collaborated in studies demonstrating that a single oral dose of the sympathetic alpha-agonist midodrine taken at the end of simulated space flight is effective in markedly reducing the development of orthostatic hypotension. Studies to test this drug in astronauts are in progress.
Robert S. Lees, professor of health sciences and technology, in collaboration with Roger Kamm, professor of mechanical engineering, and Raymond Chan, former medical engineering graduate student and currently instructor in radiology at Massachusetts General Hospital and Harvard Medical School, have developed a noninvasive method for estimating carotid arterial motion and strain from real-time ultrasound image sequences. The blood pressure variation in the carotid artery with each heartbeat causes cyclic changes in its wall, which relate directly to the amount of arteriosclerosis in the vessel. Early changes in these arteries, which supply blood to the brain, can therefore be monitored with this novel method to improve diagnosis of early carotid atherosclerosis and ultimately aid in the prevention of stroke prevention.
Daniel C. Shannon is a founder of the field of pediatric intensive care and pulmonology. For more than two decades, he has been furthering his breakthrough studies into the rare condition of congenital central hypoventilation syndrome (CCHS). Dr. Shannon and his colleagues have added incrementally to their framework of knowledge about the problem, using whatever new tool they could find to test specific hypotheses. One of his associates in these studies recently received an award from the NIH for discovering the first gene that controls respiration. Still, the clinical problems—like a child's failure to breathe adequately when asleep—continue to stump clinicians and researchers alike. His latest research reflects the use of technology to serve patients better.
Future Plans
- Promote the Athinoula A. Martinos Center as the leading imaging and research center in the country. Plans are underway for Brainstorm 2002, a neuroimaging conference in Athens, Greece, in September 2002. This will involve neuroscientists from the Martinos Center and other top neuroscientists from around the world.
- Hold the HST Symposium "Experiencing the Frontiers of Biomedical Technology on March 10 and 11, 2003. This biennial event will feature a unique opportunity for participants to experience first-hand the technologies involved in tissue engineering, drug delivery systems, human-machine systems, and informatics.
- Build on the robust health of HST's educational programs and expand them, especially in the fields of bioinformatics, biomedical enterprise, and speech and hearing science and technology.
- Expand HST's research presence. Future HST research foci will be primarily directed in three areas: biomedical imaging, biomedical informatics and computational biology, and regenerative biomedical technologies.
- Continue exploration of mechanisms to optimize impact on education and research at the interface of biomedical/bioengineering in the health sciences.
- Expand HST's faculty and its visibility in research in the Harvard and MIT communities.
More information about the Harvard-MIT Division of Health Sciences and Technology can be found on the web at http://hst.mit.edu/.