Picower Center for Learning and Memory
The Picower Center for Learning and Memory, founded in 1994, focuses on understanding the mechanisms underlying learning and memory at molecular, cellular, and brain-circuit levels. We have been successful in assembling a group of talented investigators from different backgrounds who interact synergistically in collaborative research efforts, both formally and informally, and in attaining funding for growth. In 2001, the Picower Foundation endowed the Picower Center for Learning and Memory with a gift that will fund expansion in the form of a new state-of-the-art building, four endowed Picower professorships, and endowment for research and operations. The Riken Brain Science Institute, an international center of excellence based in Saitimo, Japan, supports the Riken–MIT Neuroscience Center, which is comprised of seven investigators and has recently increased the level of funding. Another major collaborative program is the National Institute of Mental Health program in genetic, physiological, and behavioral studies of memory, which supports five faculty investigators. Picower faculty are also supported by individual research awards, predominately from the National Institutes of Health.
In academic year 2003, the center hired two new faculty members, bringing the number of total active faculty to 11. Of these, six are tenured and five are nontenured, three are Howard Hughes Medical Institute investigators, and one is a Nobel laureate. The growth in the faculty was accompanied by a commensurate growth in research staff, bringing the total number of faculty, staff, and students of Picower Center to 165. A highlight of the year was the third annual Picower–Riken Neuroscience Symposium, held at MIT in March and attended by over 300 participants.
Research Highlights
The Bear laboratory has made significant progress elucidating the synaptic and molecular bases of receptive field plasticity in the visual cortex. One example of receptive field plasticity, originally described by Wiesel and Hubel 40 years ago, is the striking loss of visual cortical responsiveness to an eye temporarily deprived of normal vision during a critical period of postnatal life. Bear has shown that this deprivation-induced synaptic depression is a consequence of residual retinal afferent activity that fails to correlate with evoked postsynaptic responses in the visual cortex. To understand the mechanism, the lab established the model of homosynaptic long-term depression (LTD) in brain slices. Recent work has now shown that a period of monocular deprivation (MD) triggers the same molecular changes in AMPA receptor function as LTD. Therefore, MD induces LTD. Bear will continue the molecular dissection of neocortical LTD, with the ultimate aim of generating conditional mouse mutants sites, to understand the relative contributions of LTD to the effects of MD, and to determine the molecular basis of the critical period.
The Hayashi laboratory focused on several projects dealing with the molecular biology of excitatory synaptic transmission. They have been working on the molecular mechanisms underlying long-term potentiation (LTP) of hippocampal CA1 synapse. They previously found that LTP induction delivers AMPA-type glutamate receptors into the synapse, which contribute to the enhanced transmission. The lab is currently working to elucidate the detailed molecular mechanism of this phenomenon by combining electrophysiology, two-photon microscopy, and molecular biology. They also found a motoneuron-specific subunit of NMDA receptor subunit NR3B and are currently testing if a dysfunction of this receptor causes motoneuron disease represented by amyotrophic lateral sclerosis (Lou Gehrig's disease).
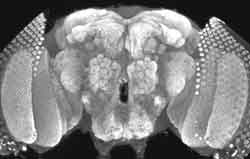 |
| Localization of synaptotagmin to Drosophila neuromuscular junctions. |
The focus of the Littleton laboratory is to elucidate the molecular mechanisms underlying synapse formation, function, and plasticity. They combine molecular biology, protein biochemistry, electrophysiology, and imaging approaches with Drosophilagenetics to investigate the molecular mechanisms involved in neuronal signaling. Using DNA microarray analysis on conditional mutants in Drosophila that induce neuronal hyperexcitation, they have analyzed the Drosophila genome for activity-regulated gene expression in the fly brain. These approaches have allowed them to identify many previously unsuspected candidates for activity-dependent modulation of neuronal function. The lab is now determining how these genes contribute to cellular forms of behavioral plasticity by analyzing their ability to modulate neuronal function or connectivity. Together, these approaches should greatly expand the understanding of the basic mechanisms of synapse function and plasticity, as well as provide insights into expression changes that allow synaptic ensembles to store information through changes in neuronal connectivity and function.
The Liu laboratory focuses on identifying the principles that guide the formation of functional neural circuits. In the past year, this lab has done a series of experiments to uncover principles governing the organization of synaptic inputs within a dendritic tree. They found that excitatory and inhibitory synapses are organized in an ordered fashion in a dendritic tree to maintain a local balance of excitation and inhibition. This synaptic organization is maintained by a "push-pull" regulatory mechanism. Surprisingly, they found that this balance of excitation and inhibition is essential for plasticity of synapses. These findings may further the understanding of how synaptic plasticity is regulated in vivo.
The Lois laboratory studies neurogenesis in the adult brain and the assembly of neuronal circuits. Retroviruses have become one of the preferred viral-based gene delivery vehicles because of their ability to permanently integrate into the genome of target cells. However, previous attempts to generate transgenic animals by delivering genes to embryos with oncoretroviruses resulted in developmental silencing and failure to express the transgene. Recently (Science 2002, 295:868) they have demonstrated that lentiviruses (another family member in the retroviridae class) are not subjected to the developmental silencing previously observed with oncoretroviruses. Furthermore, lentiviral-based vectors can be used to express genes in specific tissues or cell types when engineered with tissue-specific promoters. Because of these properties, lentiviral vectors could allow for the genetic modification of species that until now have been refractory to transgenic approaches, such as birds or nonhuman primates.
The Miller laboratory uses experimental and theoretical approaches to study the executive brain functions that organize complex thought and action. This work focuses on the prefrontal cortex, the region of the brain most elaborated in intelligent animals such as humans and other primates. Their studies have provided insight into how the prefrontal cortex and related brain areas acquire the knowledge about the high-level, abstract categories, concepts, and rules needed to guide intelligent, goal-directed behavior. These findings have provided a foundation upon which to construct more detailed, mechanistic accounts of how executive control is implemented in the brain. In the past year, the Miller Laboratory has made key discoveries about how and where concepts such as "number," "cat and dog," "same," and "different" are represented in the brain. They have also found that their representation shares fundamental properties with lower-level information, such as brightness. This suggests a continuum between lower- and higher-level brain functions and helps resolve one of the oldest and most fundamental debates in cognitive science. The lab has also provided key information on how the brain pieces together information about ongoing actions and their consequences.
The Nedivi laboratory has been working on characterizing CPG2, a gene they isolated in a forward genetic screen for activity-regulated genes that may play a role in synaptic plasticity. Its message is expressed in brain regions capable of synaptic plasticity and is regulated in the visual cortex by exposure to light. The CPG2 protein localizes to subsynaptic regions of dendritic spines and interacts with the actin cytoskeleton. To address its function at the synapse, the lab generated a lentivirus targeting vector containing a small-hairpin ribonucleic acid (shRNA) specific to CPG2. Neurons infected with the resulting lentivirus show a >80 percent decrease in CPG2 levels within individual spines. The CPG2–shRNA lentivirus will provide a valuable tool for determining the effects of reduced CPG2 levels on synaptic function.
The Sheng laboratory is interested in the molecular mechanisms by which synapses in the brain change their strength and connectivity in response to experience. In the adult brain, there has to be a long-term balance between the formation of new synapses and the loss of old (poorly used) synapses. During the past year, the Sheng lab has discovered a novel way to induce synapses (involving glutamate receptor protein GluR2) and several novel ways by which brain cells eliminate synapses. The latter studies are illuminating a new area of molecular neuroscience that has potential applicability for treatment of neurodegenerative disease.
The Sur laboratory made two important discoveries about the organization and function of the visual cortex. By combining optical imaging and intracellular recording in vivo, the lab showed that neurons at specific locations—pinwheel centers—integrate a wide range of inputs and are thus particularly capable of plasticity in their responses. By recording monkeys performing visual fixation tasks, the lab demonstrated that neurons at the earliest stages of visual cortical processing can convey information about internal cognitive representations, such as judgments of stimulus probability that regulate where we look next.
The Tonegawa laboratory made major discoveries in two areas. Using a novel genetic technology developed in this lab, researchers have created mouse strains in which only one of about 30,000 mouse genes—and, therefore, the protein it creates—is "knocked out," only in a particular type of neuron of a highly restricted part of the brain. By observing the physiological and behavioral deficits these mice display, Tonegawa's team, in collaboration with Matt Wilson's lab, discovered that a single gene encoding a neurotransmitter receptor, called NMDA receptor, in the specific tiny area of the hippocampus called area CA3, is critical for two major memory functions: the ability to rapidly form memories of one-time experiences and to recall the details of the memory previously formed with scant information as the recalling cues.
In the second major work, Tonegawa's laboratory knocked out another gene for an enzyme calcineurin only in the front part of the brain. This mouse strain displayed a number of behavioral deficits shared by human schizophrenia patients, such as reduced "working memory" (the memory needed, for example, to remember where you placed your reading glasses a few minutes ago), impaired attention, and diminished social interactions. Tonegawa and his collaborators further showed that variation in a human calcineurin gene is indeed associated with schizophrenia. This is the first study that uses animals demonstrating an array of symptoms observed in schizophrenia patients to identify specific genes that predispose people to the disease. The work provides novel molecular targets for the development of new therapeutic and diagnostic methods for schizophrenia and possibly even for other related psychiatric diseases such as bipolar disease and autism.
The Wilson laboratory has continued to focus on the role of the hippocampus and its interactions with the neocortex during sleep and waking states in the formation and maintenance of memory in the mammalian nervous system. Recently published work in collaboration with Susumu Tonegawa has demonstrated for the first time the role of the circuits within hippocampal area CA3 in the formation of memories of novel events that may relate to the formation of human memories of personal experience (Nakazawa et al.). The lab has also published work demonstrating the replay of memories for sequences of events during slow-wave sleep in a study that has extended our understanding of the role of sleep in the establishment of long-term memories of experience (Lee and Wilson).
Faculty Changes
Two exciting faculty appointments have been made during the last year. Carlos Lois, who studies neurogenesis in the adult brain and the assembly of neuronal circuits, was hired as assistant professor. Mark Bear, who seeks to understand how synapses in the cerebral cortex are modified by experience, was hired as professor. The center plans to hire two more faculty members over the next couple of years, bringing the total number of Picower laboratories to thirteen.
Honors and Awards
Mark Bear, who was appointed Picower professor in 2003, gave the Chancellor's Award Lecture in Neuroscience at the Neuroscience Center of Excellence at Louisiana State University and the Grass Traveling Scientist Lecture at the University of Pittsburgh. Yasunori Hayashi received the Ellison Medical Foundation New Scholar in Aging Award. Troy Littleton was awarded a Packard Foundation Fellowship. Carlos Lois received a Poitras professorship. Earl Miller was named editor, Journal of Cognitive Neuroscience, was elected to the International Society for Behavioral Neuroscience, and gave the keynote address at several major conferences. Elly Nedivi was awarded the Dean's Education and Student Advising Award. Mriganka Sur was elected fellow of the American Academy of Arts and Sciences. Susumu Tonegawa was invited to give the 2003 Chancellor's Award Lecture in Neuroscience at the Neuroscience Center of Excellence at Louisiana State University, the Cartwright Award Lecture at Columbia University College of Physicians and Surgeons, and the Presidential Lecture at the Society for Neuroscience annual meeting in 2002.
More information about the Picower Center for Learning and Memory can be found on the web at http://mit.edu/picowercenter/.