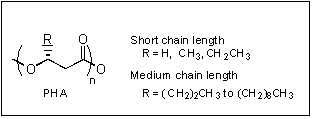

Polyhydroxyalkanoates (PHAs) are polyoxoesters that are produced by a wide range of bacteria as energy storage compounds (Fig. 1). PHAs have properties ranging from thermoplastics to elastomers, making them useful for the production of bulk and specialty plastics. PHAs are also biodegradable. Given these features, PHAs are interesting in the newly emerging field of biopolymer engineering, a major new application for recombinant DNA technology. My lab, in recent collaboration with Prof. JoAnne Stubbe in the Dept. of Chemistry, has been focused on understanding the production of PHAs as this process occurs in vivo. We have been studying the role of two classes of proteins, PHA synthases and phasins, in PHA production. Recently, my lab has entered into an interdisciplinary partnership with Malaysian institutions and programs to study the microbial production of PHAs using palm oil, and other natural compounds derived from oil palm, as a carbon source. Our long-term goal is to develop rational strategies for the controlled synthesis of PHA biopolymers.

Fig. 1. Structure of PHAs.
PHA synthases play the central catalytic role in PHA synthesis and granule formation by catalyzing the polymerization of hydroxyacyl-CoA substrates to yield PHAs, which in turn associate to form PHA granules. Our studies of the PHA synthases of Ralstonia eutropha (PhaCRe) and Allochromatium vinosum (PhaECAv) have yielded important insights on the mechanism of PHA synthesis, namely that a cysteine residue conserved among these two proteins (PhaCRe C319 and PhaECAv C149) and all known PHA synthases is involved in covalent catalysis and that the synthases share structural and functional similarities with lipases.
Previously, we have focused on comparing and contrasting the properties of PhaCRe and PhaECAv, particularly with regard to enzyme mechanism and substrate specificity. Regarding mechanism, we have constructed a set of PhaCRe and PhaECAv mutants with substitutions at conserved amino acid residues and we have tested the activity and PHA producing abilities of these mutant proteins in vitro and in vivo. Along with C149 (A. vinosum numbering), H331 and D302 were found to be crucial for catalysis. We previously suggested that the first two residues work as a catalytic dyad, in which H331 activates C149 for nucleophilic attack on 3-hydroxybutyryl-CoA (HBCoA, the enzyme's substrate), while D302 is proposed to function as a general base catalyst to activate the hydroxyl group of HBCoA for ester formation. Regarding substrate specificity, we have determined that PHA synthase can recognize, in addition to the normal substrate R-3-hydroxybutyryl-CoA, two substrate analogs, lactoyl-CoA and R-3-hydroxybutyryl-N-acetylcysteamine. These observations suggest potential applications for PHA synthases in producing polylactides in vivo and PHAs in vitro.
In contrast to PHA synthases, phasins play a poorly understood role in promoting PHA synthesis and granule formation. Phasins are low molecular weight proteins, designated PhaP, that share no sequence homology and that have been identified from many bacterial strains based on their accumulation to high levels in cells producing PHAs and their association with PHA granules. Phasins from several bacterial strains have been shown to increase production of PHAs and to promote the accumulation of PHAs as numerous small granules in cells. These two effects are likely related but the precise role played by phasins remains to be determined. However, we have constructed an R. eutropha strain deleted for the phaP gene (DphaP) and have compared production of PHA by the wild-type and DphaP strains across a time course and under a range of cultivation conditions. Our results indicate that PhaP must accumulate to high levels in order to promote PHA synthesis and suggest possible roles for PhaP.
To refine understanding of regulation of phasins, we have also focused on the role of a specific protein, PhaR, in regulation of PhaP accumulation. PhaR is a DNA binding protein that has been proposed to play a role in regulating PhaP accumulation based on the proximity of the phaR and phaC genes and based on analysis of phasin regulation in heterologous systems. We have conducted genetic analyses in R. eutropha and Escherichia coli that demonstrate that PhaR is a negative regulator that couples PhaP accumulation to PHA production in cells. Importantly, PhaR/PhaP regulation can be reconstituted in heterologous systems, implying that PhaR likely recognizes PHA and regulates the phaP gene directly. Indeed, experiments conducted in our lab indicate that PhaR does bind to a sequence upstream of the phaP gene, and a set of mutations engineered in the phaR gene is beginning to reveal which amino acid residues are crucial for DNA binding and PHA response.
All biochemical characterization of PhaCRe performed to date has been performed on the recombinant enzyme, expressed in E. coli. Complete biochemical characterization of PhaCRe has proved difficult, due to the presence of a lag phase in enzyme activity of the recombinant synthase. Recently, we have recovered a highly purified, epitope-tagged PhaC from R. eutropha and attempted biochemical characterization of the native enzyme. No lag phase in activity was seen from this native enzyme, allowing us to also obtain accurate Km and Vmax values for the first time. We are currently exploring the nature of the lag phase in the recombinant enzyme.