Karishma Rahman
Massachusetts Institute of Technology
Ting Laboratory
Mentor: Amar Thyagarajan
Summer 2008
Developing an enzyme labeling based methodology to image protein-protein interactions at live neuronal synapses
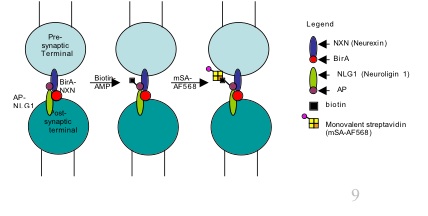
In the nervous system, information is exchanged at neuronal synapses. Neurexin (NXN) and neuroligin (NLG) are proteins found on the surfaces of pre and post-synaptic terminals respectively. The NXN-NLG interaction complex induces the clustering of proteins normally present at excitatory or inhibitory postsynaptic sites, however, when during the course of synaptogenesis, this interaction occurs is unclear primarily, because of lack of a suitable method to image this phenomenon. Current methods to study such protein-protein interactions are limited to colocalization assays using fluorescent protein fusions or antibody staining. However, such methods are constrained by the limit of resolution of optical diffraction (~200nm). Thus when two proteins are deemed to colocalize using optical microscopy methods, all one can say with certainty is that they are within a 200 nm distance of each other. In addition many proteins (such as neurexin) do not tolerate fusion of fluorescent protein tags and the tags often create problems in protein trafficking. The Ting lab has developed an enzymatic labeling based methodology that allows the imaging of NLG1-NXN interactions at excitatory synapses. In this method, the E. coli biotin ligase (BirA) is fused to NLG1 and its substrate the acceptor peptide (AP) is fused to NXN. When NLG and NXN interact at synapses and biotin is added, BirA ligates the biotin onto the AP which is then detected via a strepatavidin-AlexaFluor conjugate. This provides a direct read out of the NXN-NLG interaction at synapses. To develop a complementary system, we wanted to design a BirA-NXN/AP-NLG1 pair (opposite orientation of BirA and AP). For this purpose, I had to create a BirA-NXN fusion construct which would traffick properly to axons. I then demonstrate that the BirA-NXN and AP-NLG1 can also provide a readout of NXN-NLG interaction at the cell surface demonstrating the versatility of this enzymatic labeling method.