Samuel Thompson
Massachusetts Institute of Technology
Ting Lab
Mentors: Dr. Justin Cohen and Dr. Sujiet Puthenveetil
Summer 2009
Engineering Fluorophore Ligases for Live Cell Imaging
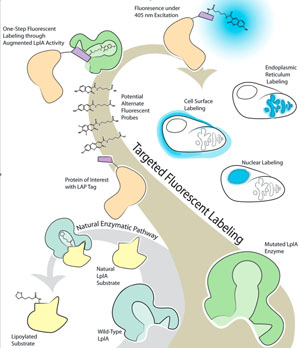
The Lipoic Acid Ligase (LplA) enzyme of Escherichia coli covalently ligates the cofactor lipoic acid to a specific lysine residue on each of its three natural substrates. This substrate specificity and the plasticity of LplA's small molecule binding pocket render LplA a favorable candidate upon which to develop a fluorophore ligation system for targeted protein imaging. It has been shown that E.coli LplA can be mutated to ligate 7-hydroxycoumarin onto both its natural substrate and an evolved 13-mer peptide tag with high specificity. Despite this initial success, previously examined mutants show little or no ligation activity for many desirable fluorescent probes. Our current endeavor is to expand the library of rationally designed fluorophore ligases to further examine known malleable residues and characterize promising new positions for rational mutation by screening new mutants for activity against a panel of such probes as Pacific Blue coumarin, 7-aminocoumarin, and resorufin. For this assay, crude cell lysate containing a single variant of mutated LplA is incubated in a reaction mixture containing the natural substrate E2p and a fluorescent probe. Ligation is confirmed by UHPLC/MS. Active mutants will be further characterized to determine the both most active mutants and trends corresponding to increased activity.