Sarah Whiteside
Massachusetts Institute of Technology
Drennan Lab
Mentor: Yan Kung
Summer 2009
Alteration of SyrB2 Halogenase Activity via Site-Directed Mutagenesis
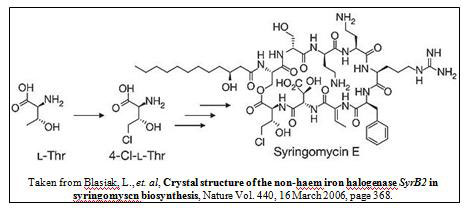
Syringomycin biosynthetic enzyme II (SyrB2), a key protein involved in syringomycin biosynthesis, is a non-heme iron enzyme that is responsible for converting L-Thr to 4-Cl-Thr. This chlorination reaction is an essential step in the synthesis of this antifungal agent. The goal of this work is to create SyrB2 mutants which can be biochemically and structurally characterized using x-ray crystallography as a primary tool. Specifically, this work aims to investigate the halogen specificity and reactivity of the enzyme. We used site-directed mutagenesis to alter the primary and secondary sphere of the Fe(II) cofactor in the enzyme's active site, most importantly around the halogen binding site near residue A118. The differing active site environments may allow other halogenation reactions, such as di- or tri-chlorination. While the basis for performing multiple halogenations over single halogenations has not yet been elucidated, these mutations, based on the sequence of barbamide biosynthesis enzymes I and II (which can trichlorinate leucine), may allow SyrB2 to catalyze multiple chlorination reactions on a single substrate, providing clues as to how the reactivity is controlled. Also, by enlarging the binding pocket, we hope to allow iodine binding in the active site, which has not yet been seen. If iodine can bind in the pocket, it should allow the iodination reaction, an unknown reaction for this class of enzymes. Overall, by mutating the primary enzyme sequence, we aim to create new enzymes that will not only tell us about how nature controls its use of halogens but can also aid in the production of biosynthetic natural products. This research has the potential to enable custom-designed compounds which can be medicinally useful.