 |
 |
A Study into the Molecular Origins of Biocompatibility :
Intermolecular Interactions between Human Serum Albumin and
Various Chemically Modified Surfaces Studied via High Resolution Force Spectroscopy
MONICA A. RIXMAN and CELIA
E. MACIAS
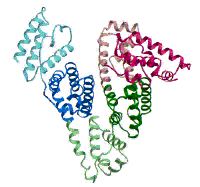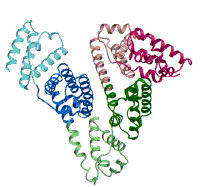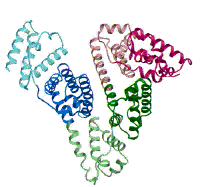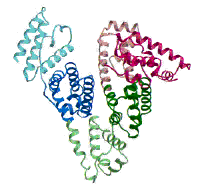
(Figure adapted from [1])
I.
Background
Polymeric coatings in the form of physisorbed or end-grafted "brushes" are frequently employed to improve the biocompatibility of a wide array of biomaterials. One of the greatest challenges to biomaterials scientists is preventing the nonspecific, noncovalent surface adsorption of proteins; the well-known first step in many undesirable processes such as triggering inflammation around artificial organs, fouling of contact lenses, and accelerated clearing of bare liposomes by the reticuloendothelial system [2]. The adsorption process is determined by the total effective interaction free energy between the protein and the surface, U(D) (where : D is the protein-surface separation distance), which is a sum of the protein-brush and protein-substrate interactions. U(D) will have various repulsive and attractive contributions that lead to complicated intermolecular potentials whose shape varies with the strength and range of the constituent interactions[3,4].
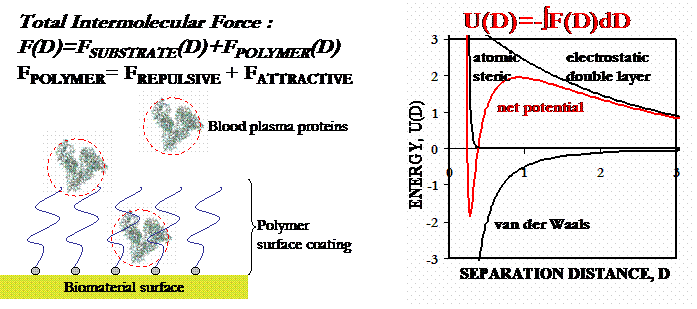
To produce protein-resistant biomaterial surfaces, it is clear that one must
design systems where repulsive interactions are maximized and attractive ones
are minimized. Steric interactions are of prime importance for end-grafted
polymer brushes (s greater than 2RF
and Lo greater than RF, where:
s is the distance betwen polymer grafting sites on the substrate, RF
is the Flory radius of an individual polymer chain, and Lo
is the height of the polymer brush). In addition, the compression of the brush
is opposed by a strong hydrodynamic lubrication force due to lateral
expulsion of the solvent residing between the protein and the surface, and
excluded volume interactions will lead to an osmotic penalty since
the protein volume becomes inaccessible to the polymer monomers.

For
compression of low density surfaces of isolated, end-grafted polymer "mushrooms"
(s
less than 2RF), the steric force arises from a free energy
penalty for chain segment surface confinement, as well as an entropic
elastic penalty for chain stretching in the plane perpendicular to the compression
axis. Other repulsive forces may include; hydrophilic hydration forces,
orientational entropy penalties for rod-like polymers, enthalpic penalties for
disruption of polymeric supramolecular (secondary) structure, and electrostatic
repulsion due to overlap of the surface diffuse counterion electrical double
layers (for charged systems). Attractive protein-brush and protein-substrate
forces may include; hydrophobic interactions, hydrogen bonding, and/or van
der Waals (VDW) forces. In order to predict the mathematical form of many
of these interactions, a number of theories have taken the approach of modeling
the proteins as dense, rigid, structureless colloidal particles with nonadsorbing
surfaces and the polymer brush as simple flexible chains.
Based on our experimental results, we have also taken this approach.
The
task of designing protein-resistant biomaterial surfaces remains elusive because
the fine details of the protein-surface intermolecular interaction are poorly
understood. The objective of this project is to directly quantify the individual
constituent intermolecular interactions between proteins and individual end-grafted,
synthetic polymer chains under a variety of environmental conditions using high-resolution
force spectroscopy (HRFS). The
protein-polymer interaction force, F, versus separation distance, D, is employed
to calculate the interaction potential, U(D=-dF(D)/dD.
The initial model system employed is the neutral, flexible, water-soluble, synthetic
polymer, poly(ethylene oxide) (PEO), -[CH2-CH2-O]n-,
which is the most commonly used material for biomaterial surface coating, and
human serum albumin (HSA), the most abundant blood plasma protein in
the human body and typically the first to adsorbt o an implanted biomaterial
surface. Our well-characterized and versatile experimental system and set-up
has a number of distinct advantages, including the ability to access individual
polymer molecules, a strong end-grafting linkage via covalent attachment of
the polymer chains to the surface, control over the underlying substrate properties,
the capacity to use a wide range of polymer molecular weights, control over
s
and Lo, and the ability to easily vary the experimental
environmental conditions. In
addition, by running "control" experiments between the protein and
a polymer-free substrate, we may quantify and subtract the protein-substrate
interactions from the overall U(D) and thereby isolate the protein-polymer interactions
in the primary experiments.
Finally,
manipulation of the experimental conditions such as ionic strength, mixed solvent
conditions, and removal of bound lipids from the protein, is coupled with theoretical
models in order to evaluate, and in some cases quantify, the constituent
intermolecular contributions to the total protein-polymer interaction (e.g.
electrostatic, steric, hydrophobic, van der Waals, etc.).
By elucidating the molecular origins of the exceptional biocompatible properies
of PEO, in particular its ability to resist the nonspecific adsorption of HSA,
we
intend for the proposed studies to serve
as a framework that may enable the manipulation and control of protein-polymer
intermolecular interactions needed to prevent protein adhesion to biomaterial
surfaces.
II.
Methodology: High Resolution Force Spectroscopy
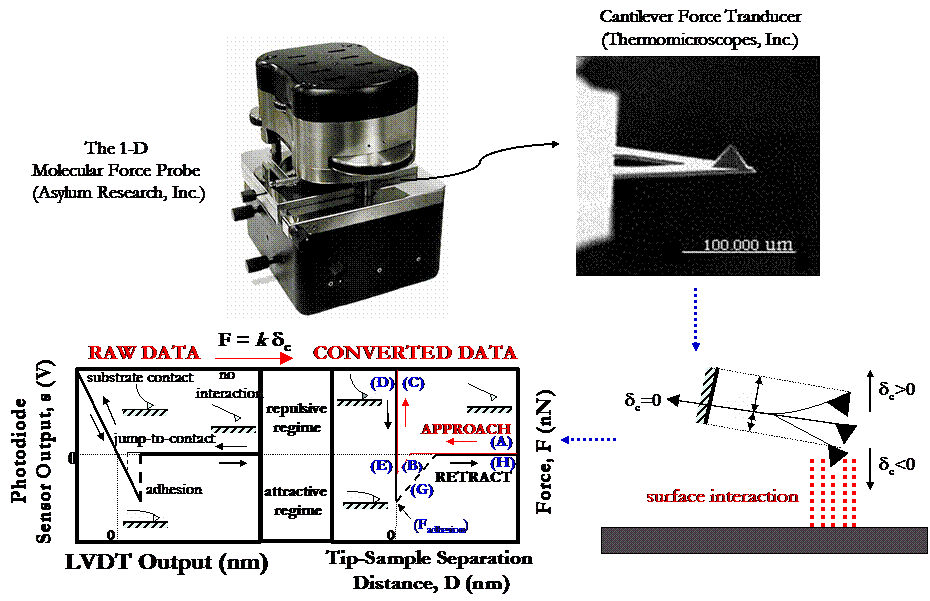
All HRFS studies were conducted using the 1-dimensional Molecular Force Probe (MFP), a new, state-of-the-art instrument produced by Asylum Research, Inc. (Santa Barbara, CA). For the very soft cantilevers used in this study (manufacted by Thermomicroscopes, Inc., and typically have spring constants k ~ 0.01 - 0.03 N/m), the MFP is able to obtain a thermal-oscillation limit of force detection of 5pN in aqueous solution. Similar to the AFM, a focused laser beam is reflected off the backside of a microfabricated cantilever force transducer with a fine probe tip into a position sensitive photodetector (PSPD). The vertical sensor output difference of the top minus bottom quadrants of the PSPD, s (nA), is used to measure the cantilever deflection, dc(nm), which is then converted into force using Hooke's Law for a simple harmonic oscillator, F = kdc (nN). In a typical HRFS experiment on a hard substrate (illustrated in the figure below), data acquisition is initiated and s (nA) is recorded as a function of z-piezo deflection, z (nm), where z is the axis normal to the sample surface that is converted into tip-surface separation distance, D (nm). The piezo incrementally moves the tip towards the sample (the "approach") in the z-direction at a constant rate. Far away from the surface (A), no interaction is present and the cantilever remains undeflected at zero force. Typically, as the probe approaches close to the surface, non-specific, attractive tip-surface interactions are present (e.g. due to van der Waals forces, etc.) and the cantilever exhibits a mechanical instability when F/D > k (the cantilever spring constant) which results in a discontinuous "jump-to-contact" (B). All data in this region of the force curve is lost, and the slope of this line is just equal to the cantilever spring constant, F/D = k. After surface contact is made, a region with an apparent infinite slope (which is set to D = 0 nm), the "constant compliance regime" (C), is observed due to the fact that the spring constant of the cantilever is much less than the stiffness of the substrate (kcantilever<<ksample). Hence, little or no deformation of the sample takes place and the cantilever and sample move together in unison. When the desired maximum repulsive force of a few nN is achieved, the piezo reverses direction and the tip is moved away from the sample ("retract") (D). Surface adhesion is usually present (E) causing the cantilever to suddenly "pull-off" from the surface (F), exhibit the same mechanical instability as observed on approach (G), and finally return to its undeflected position (H). The value of the force at the pull-off point is referred to as the adhesion force, Fadhesion. Generally, hundreds of force versus distance curves are taken for each of the experimental conditions in order to obtain a reasonable statistical representation of the data. In our experiments, the total distance, D, that the piezo travels from the start of an experiment where it is totally contracted, to the point of maximum extension, is kept constant at 1 mm, and the velocity at which it travels is also kept constant at v = 1 mm/s.
III. Intermolecular Interactions Between Human Serum Albumin and Alkanethiol Self-Assembled Monolayers
HRFS is employed to measure the intermolecular interaction potential between a covalently bound HSA monolayer and self-assembled monolayers (SAMs) of n-mercapto undecanoic acid (SH-C10H20-COOH) and dodecanethiol (SH-C12H25) in aqueous sodium phosphate buffer solution (PBS), as shown below. Experiments were also conducted on unmodified gold substrates, as a control. The objective of this work was to characterize the nanomechanical behavior of the proteins on the probe tip in relation to their hydrophobic character (with the R-CH3 SAM surface), as well as their electrostatic and hydrophilic character (on the R-COO- SAM surface).

HRFS experimental setup of an HSA-grafted probe tip vs an alkanethiol self-assembled monolayer, in sodium phosphate buffer solution (PBS)
Si3N4 cantilever probe tips were covalently coated with HSA using the methods described by Vansteenkiste, et al.[5] The success of this procedure was verified by atomic force microscopy, contact angle measurements, and fluorescence microscopy, as well as HRFS studies in poor solvent conditions. The radii of curvature of our modified probe tips have been determined by scanning electron microscopy (SEM) to be ~50-100nm. Depending on the assumed geometry of the protein,[6-8] anywhere from 68-580 proteins are within the interaction area during each approach-retract cycle of a force curve. The details of this calculation, well as the probe tip and substrate chemical modification procedures, can be found in our published works.[9]

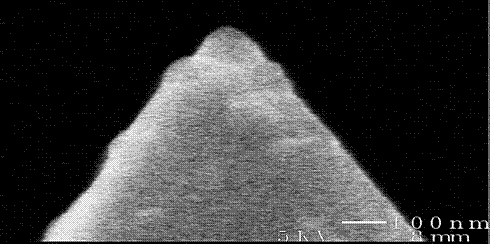
Left: Fluorescence micrograph of an HSA-grafted cantilever, taken with the assistance of Professor Darrell Irvine (MIT Department of Materials Science and Engineering). The protein preferentially binds to the probe tip, perhaps due to its geometry. Right: SEM image of an HSA-grafted cantilever probe tip: Rc ~ 65 nm.
Results on Approach
The approach to both the R-COO- and Au surfaces was purely repulsive, and can be fit well to numerical models using the nonlinear Poisson-Boltzmann theory for electrostatic repulsion in the presence of divalent ions,[10,11,12] as is shown in the figure below.
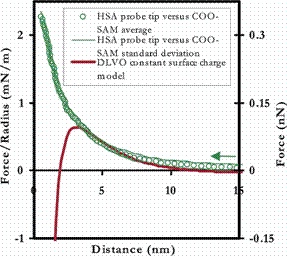
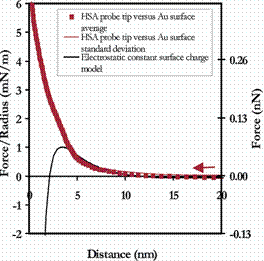
Averaged approach curves of an HSA-grafted cantilever vs a COO- -terminated SAM (Left) and unmodified polygranular Au (Right) in PBS (with standard deviations), compared to electrostatic models
In contrast, on approach of the HSA-grafted probe tip to the hydrophobic R-CH3 surface, the interaction potential was purely long-ranged attractive at D<80nm, with a jump-to-contact at D<20nm (shown below). These observations are consistent with previous observations of long-ranged attractive forces between hydrophobic surfaces.[13-15]
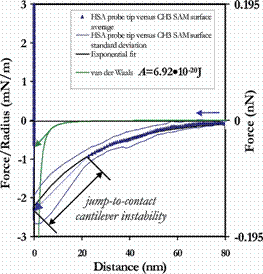
Averaged approach curves of an HSA-grafted cantilever vs a CH3 -terminated SAM in PBS (with standard deviations), compared to theoretical models for van der Waals and long-ranged, hydrophobic interactions
Results on Retract
On retract, the surfaces showed great variety in their adhesion behavior. The R-CH3 and Au surfaces exhibited the greatest forces of adhesion, and their primary mechanism was simple surface adhesion, with occasional protein extension. The strength of the surface adhesion is so great that when the tip does separate from the surface, nearly all mechanical details are lost in the cantilever instability. Hence, it is possible that protein extension may be occuring with much greater frequency than is readily apparent. Example force curves for the R-CH3 and Au surfaces are shown below.
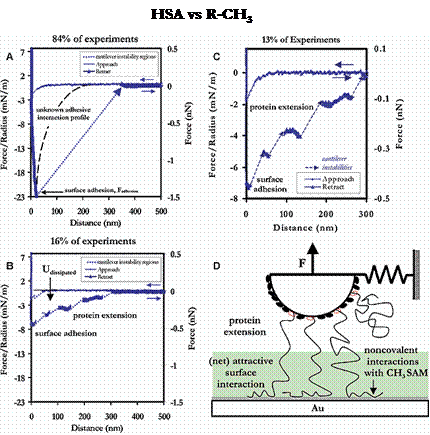
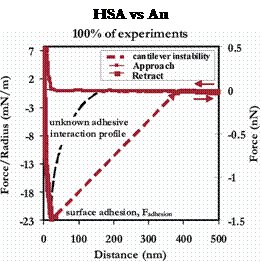
Example force curves and their relative frequencies of occurance for an HSA-grafted probe tip vs a CH3-terminated SAM (Left) and unmodified polygranular Au (Right), in PBS
The R-COO- surface exhibited a number of adhesive mechanisms as well, although half of the total force curves taken were completely nonadhesive and nonhysteretic. As with the R-CH3 and Au substrates, of the adhesion events that were observed, surface adhesion was much more common than protein extension. The average forces and distances of adhesion for these substrates is given in the following section.
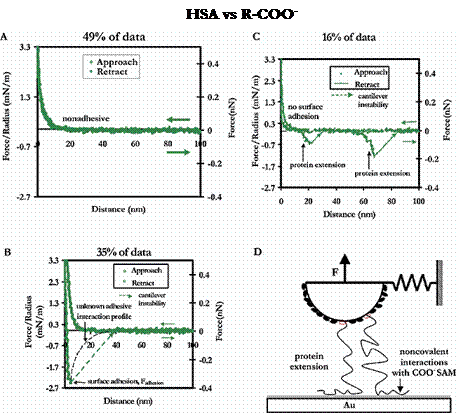
Example force curves and their relative frequencies of occurance for an HSA-grafted probe tip vs a COO- -terminated SAM in PBS
This work was recently published in Langmuir 2003, 19 (15), 6202-6218, and is available to download in *.pdf format here.
IV.
Intermolecular Interactions Between Human Serum Albumin and Individual End-Grafted
Polymer Chains
IV.A.
Experimentation in phosphate buffer solution (IS = 0.01M, pH = 7.4)
HRFS
is employed to measure the intermolecular interaction potential between a covalently
bound HSA monolayer and an individual end-grafted PEO chain (i.e. "mushroom")
in aqueous sodium phosphate buffer solution (PBS) under various environmental
conditions (e.g. ionic strength, pH, etc.) as shown below. An unmodified, polygranular
Au substrate was used as a control for this experiment.
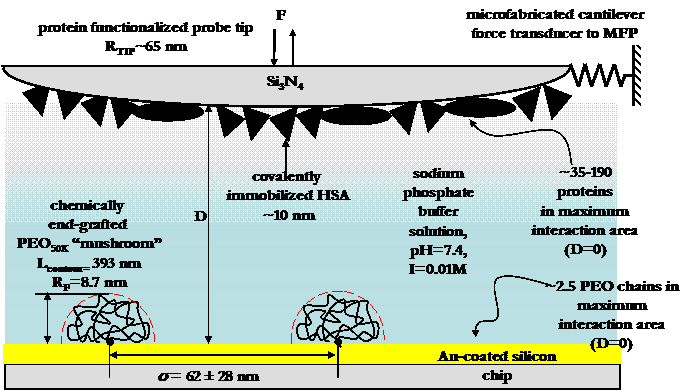
Experimental setup of HRFS experiments between and HSbA-grafted probe tip and a PEO-grafted polygranular gold surface.
Si3N4 cantilever probe tips were
covalently coated with HSA as described in the previous section. Chemisorption
of n-mercapto PEO is employed to end-graft one end of the polymer chain to a
gold substrate. Sufficiently low chain grafting densities (s
greater than 2RF, Rtip) are achieved
by reducing the chemisorption incubation time and/or concentration of polymer
in solution. Surface characterization and verification of the low polymer chain
grafting density is confirmed using atomic force microscopy (AFM) imaging and
single molecule force spectroscopy, and was found to be
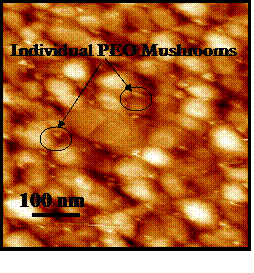
Contact mode AFM image in PBS of individual PEO mushrooms end-grafted to a polygranular gold substrate: s ~ 62±28 nm.
Results on Approach
On approach of an HSA-grafted probe tip to a polymer-grafted surface, a significant intermolecular repulsion exists which is far greater in range (beginning at D=30-50nm) and magnitude (1-3 mN/m) than can be explained by theoretical models of any one of steric, electrostatic, or van der Waals forces (as shown in the figure below), and indeed far greater even than all of these forces combined. Note that PEO is able to exhibit this large repulsion even at very low grafting densities! We believe this repulsive force to be a direct result of PEO's exceptional ability to hydrogen bond extensively with surrounding water molecules in an intricate and elegant way; for example, PEO is largely believed to form a local (11/2) helical supramolecular structure due to intramolecular H-bonds (as shown below), and intermolecular H-bonds have been observed to nucleate a new phase of water in the polymer's immediate vicinity that is closer in density to ice than to liquid water. This network of H-bonding results in a large enthalpic penalty to perturbation and disruption, which we suspect is primarily responsible for the large repulsive forces observed on approach. When identical experiments were conducted in the organic solvent hexadecane (a poor solvent for PEO), we and others [15] have found the repulsion to be eliminated and replaced by long-ranged attraction.
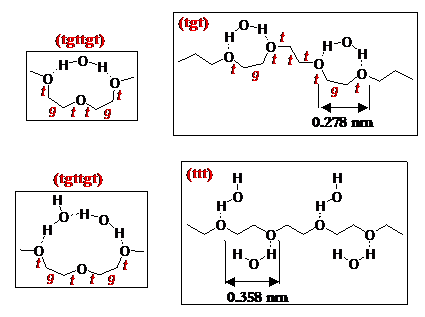
Above: Proposed mechanisms of some supramolecular structures that may form in PEO as a result of intramolecular hydrogen bonding.
Below: Averaged force profile of an HSA-grafted probe tip approaching a PEO-grafted polygranular gold substrate in PBS (IS = 0.01M, pH = 7.4). 94% (i.e. the percentage of the Au surface that is not covered by PEO mushrooms) of the HSA vs Au averaged interaction potential has been subtracted from the HSA vs PEO-Au average to yield the curve below. The observed repulsive force is greater both in range and magnitude than theoretical models for intermolecular forces, individually or combined.
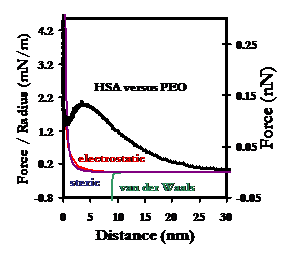
Results on Retract
On
contact of the HSA probe tip with the polymer mushrooms, the intermolecular
force becomes attractive, and complexation enables tethering and extension
of either or both the protein and polymer chains upon retract. The average forces
and distances at which complexation was broken and the tip and polymer separated
were found to be

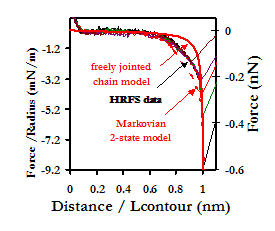
Typical retraction curve of an HSA-grafted probe tip from a PEO-grafted Au substrate in PBS, showing the extension of a single polymer chain. Comparison is made to theoretical models used to describe the extensional behavior of polymers in solution.
IV.B. Comparison of HSA vs PEO Data to HSA vs Alkanethiol SAMs
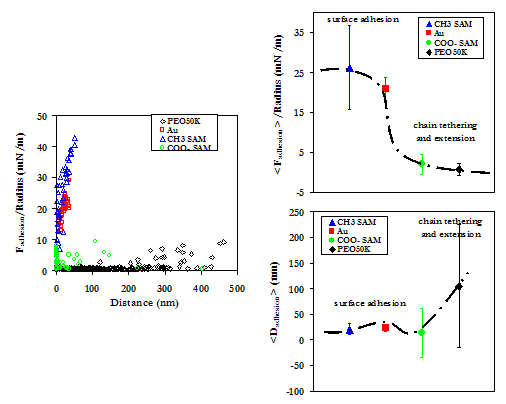
Below is a comparison of the experimental data on the PEO-grafted substrate to the alkanethiol SAMs and Au. It is clear that PEO not only is more repulsive at long range than each of the other substrates studied, but also adheres HSA with the least force. This behavior is indicative of the exceptional ability of PEO to resist the nonspecific adsorption on blood plasma proteins and the associated thrombic response.
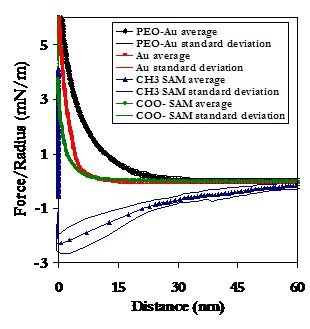
Averaged approach curves (Above), as well as the experimental data points of maximum adhesion (Below, Left), and their averages with standard deviations (Below, Right), of the HSA vs PEO data and the HSA vs Alkanethiol SAMs and unmodified Au.
This work has been accepted for publication in Langmuir, 2003, and is available to download in *.pdf format here.
IV.C. Deconvoluding the Intermolecular Interaction Potential: Varying Experimental Conditions
In order to deconvolude the total interaction potential into constitent intermolecular forces (e.g. electrostatic, steric, and hydrophobic interactions), we studied the HSA vs PEO system under a variety of experimental conditions, including three orders of magnitude of ionic strength from IS = 0.01 to 1.0M, several cantilever spring constants and rates of pulling, using lipid-bound and lipid-free HSA, and employing mixed solvent conditions, including the use of the antihydrophobic agent isopropanol.
~The following data is currently being prepared for submission for publication.~
IV.C.1. Varying Solution Conditions: Ionic Strength
Results on Approach
Two intuitive sources of the long-ranged repulsion observed on the PEO-grafted substrate are electrostatics and steric effects. Although PEO is largely believed to be a neutral molecule, an effective charge can be induced by the approaching negatively charged HSA-grafted probe tip, thus resulting in a repulsion of like-charges. In terms of steric forces, it is believed that a flexible macromolecule such as PEO spends a significant amount of time sampling conformations and dimensions up to two are three times it radius of gyration, Rg. It has therefore been often postulated that the long-ranged repulsion is due to steric hindrance of the more expanded polymer conformations.
When electrostatic repulsive interactions represent a significant contribution to U(D), increases in the ionic strength of the solution are expected to decrease the repulsive force observed on approach, as counterions from the buffer "shield" the effective charge on the surfaces of both the probe tip and the underlying substrate. Furthermore, NaCl, the dominant salt in our buffer, is believed to have a "salting out" effect on PEO, thus resulting in a decrease in Rg. Our results (shown below) do not follow either of these predictions, however, as an increase in the ionic strength of the PBS to 1.0M (by adding NaCl) results in an increase in repulsion on approach. While we currently cannot explain the origins of this phenomenon, we do conclude that neither electrostatic nor steric repulsion are significant forces in the HSA-PEO intermolecular interaction potential.
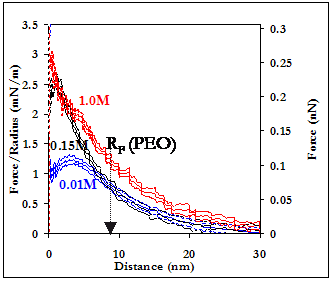
Averaged force curves of an HSA-grafted probe tip approaching a PEO-grafted polygranular gold substrate in PBS, as a function of ionic strength. 94% (i.e. the percentage of the Au surface that is not covered by PEO mushrooms) of the HSA vs Au averaged interaction potentials have been subtracted from the HSA vs PEO-Au averages to yield the curves shown.
Results on Retract
We did not observe any effect on the type, strength, or range of adhesive interactions on retract as a function of ionic strength.
IV.C.2. Varying Solution Conditions: Antihydrophobic Agents
The addition of small quantities of "antihydrophobic" agents such as isopropanol have been shown [16] to completely eliminate the presence of hydrophobic interactions by solvating hydrophobic moieties in water, as shown in the figure below.
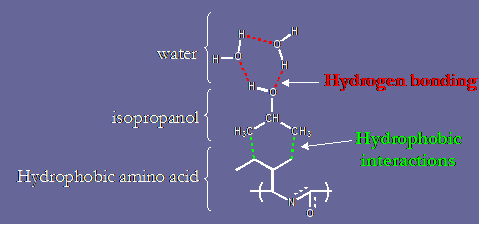
The "antihydrophobic" mechanism of isopropanol.
Results on Approach
In an effort to quantify the hydrophobic component of U(D) for the HSA vs PEO system, we studied the effect of adding 0.5% and 5% (v/v) to 0.01M PBS, as well as the effect of changing the solvent completely to 100% isopropanol. As the hydrophobic regions of HSA become solubilized in the aqueous PBS by the isopropanol, the protein is rendered completely hydrophilic. On approach of the hydrophilic probe tip to the hydrophilic PEO-grafted substrate, a repulsive interaction is expected due to competing hydrogen bonding with water molecules, as well as enthalpic penalties to the disruption of the hydrogen bonded water network within and surrounding the two surfaces. As shown below, we observed an increase in the repulsive force on approach upon the addition of just 0.5% isopropanol, the magnitude of which may be quantified as the hydrophobic force component to U(D). In 100% isopropanol (which is a poor solvent for PEO at STP), the interaction potential becomes completely attractive, possibly due to a solvophobic effect from the PEO. Although hydrophobic forces are clearly significant in the HSA-PEO intermolecular interaction potential, they are still not sufficient to explain the range and magnitude of the observed repulsive force.
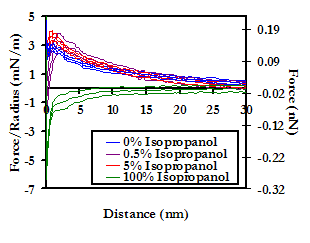
Averaged force profile of an HSA-grafted probe tip approaching a PEO-grafted polygranular gold substrate in PBS (IS = 0.01M, pH = 7.4), as a function of the concentration (v/v) of isopropanol. 94% (i.e. the percentage of the Au surface that is not covered by PEO mushrooms) of the HSA vs Au averaged interaction potentials have been subtracted from the HSA vs PEO-Au averages to yield the curves above
Results on Retract
While no effect was observed on the average forces and distances of maximum adhesion upon the addition of small amounts of isopropanol, the frequency of adhesion events showed a marked decrease as the concentration of isopropanol was increased from 0% to 0.5% (v/v). From 0.5% to 5.0% isopropanol the frequency of surface adhesion increased again slightly, but the frequency of protein pulling continued to decrease. These results (shown below) suggest that hydrogen bonding is a significant factor in the abililty of PEO and HSA to form attractive interactions while in contact. In 100% isopropanol, the frequency of adhesion increases dramatically. This result further supports the postulate presented above that the attraction between HSA and PEO in 100% isopropanol is predominantly a solvophobic effect.

Above: Experimental data points of maximum adhesion for an HSA-grafted probe tip on a PEO-grafted substrate as a function of concentration of the antihydrophobic agent isopropanol in PBS
Below: Frequencies of the mode of adhesion for the data points presented above
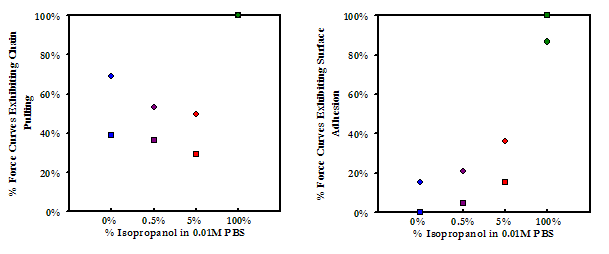
IV.C.3. Investigating the Role of Fatty Acids Bound in the Hydrophobic Pockets of HSA
It is interesting that we observed such a clear contribution from hydrophobic forces, particularly since native HSA contains on average 1-2 bound fatty acids per molecule,[17] the role of which is to sit within the hydrophobic pockets of the protein and shield them from unfavorable interactions with the aqueous environment. These fatty acids may indeed be a significant factor in why HSA is so very water-soluble. Indeed, the question arises as to what might the interaction potential be in the absence of the bound lipids, and what would the effect of antihydrophobic agents be then? To answer this question, we obtained HSA that had been stripped of 99% of its bound lipids, and repeated the experiments varying the concentration of isopropanol.
Results on Approach
The removal of bound lipids exposes the hydrophobic pockets of the protein, thus rendering it overall more hydrophobic than in the native state. It was expected, therefore, that the addition of an antihydrophobic agent and the consequential increase in hydrophilicity of the protein would result in an even greater relative increase in repulsion on approach. On the contrary, there is a dramatic reduction in the magnitude of the observed repulsion on approach upon the addition of just 0.5% isopropanol to PBS. There is no further reduction as the concentration is increased to 5%, but at 100% isopropanol the interaction potential again becomes completely attractive. We are still in the process of interpreting these results, which are shown below.
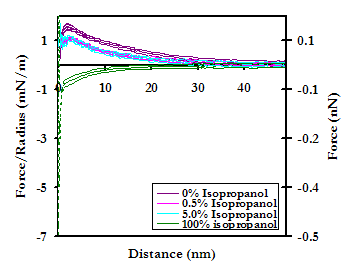
Averaged force profile of a lipid-free HSA-grafted probe tip approaching a PEO-grafted polygranular gold substrate in PBS (IS = 0.01M, pH = 7.4), as a function of the concentration (v/v) of isopropanol. 94% (i.e. the percentage of the Au surface that is not covered by PEO mushrooms) of the HSA vs Au averaged interaction potentials have been subtracted from the HSA vs PEO-Au averages to yield the curves above.
Results on Retract
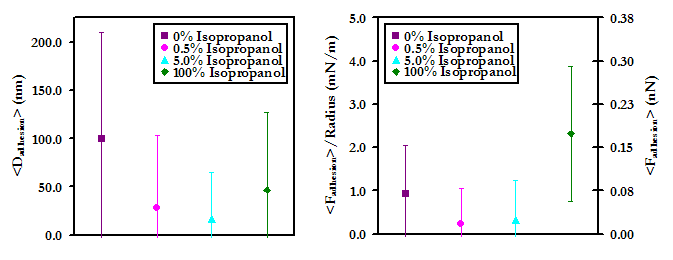
Upon the addition of isopropanol, the average forces and distances of adhesion (shown below) are seen to decrease, suggesting that hydrophobic interactions may also play a significant role in the nonspecific binding of HSA to PEO in contact. This result may possibly be explained by the fact the PEO is an amphiphilic molecule. Much like with the lipid-bound probe tip, as the solution is changed to 100% isopropanol, the forces and distances again increase, likely for the same reasons discussed previously for this solvent condition. Again much as with the lipid-bound tip, the frequency of adhesion events showed a marked decrease as the concentration of isopropanol was increased from 0% to 0.5% (v/v); from 0.5% to 5.0% isopropanol the frequency of surface adhesion increased again slightly, but the frequency of protein pulling continued to decrease; and in 100% isopropanol, the frequency of adhesion increases dramatically. These results are also shown below.
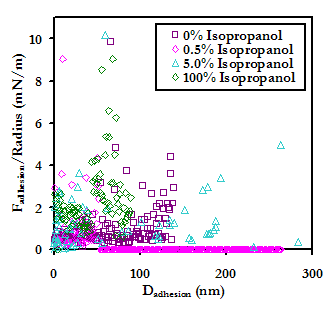
Experimental data points (Above) and average forces and distances (Below) of maximum adhesion for an HSA-grafted probe tip on a PEO-grafted substrate as a function of concentration of the antihydrophobic agent isopropanol in PBS
Below: Frequencies of the mode of adhesion for the data points presented above

IV.C.4. Investigating the Role of Tip Compliance and Scan Rate
Two additional factors that may affect the binding of HSA to PEO are related to cantilever parameters: the scan rate and the tip compliance. The former is a culprit predominantly because the formation of nonspecific attractive and repulsive interactions is a time-dependent process. The latter may play a role in the amount or degree of detail that we are able obtain from a given force curve. To explore these possibilities, we have conducted experiments to explore the effect of these two on the total intermolecular interaction potential.
Results on Approach
No effect was observed on the approach force profile as a function of tip compliance over a range of spring constants from 0.01 to 0.1 N/m.
The effect of scan rate was studied over a range from 0.75 mm/s to 6.0 mm/s, and while no significant difference was observed between 1.5mm/s and 6.0mm/s, a significant decrease in the repulsion was observed as the rate was decreased to 0.75mm/s. It is difficult to interpret this data, as classical theories on hydrodynamic effects do not hold for our experiments due to the relatively slow scan rate limit of the instrument, as well as the extremely small surface area of the probe tip.
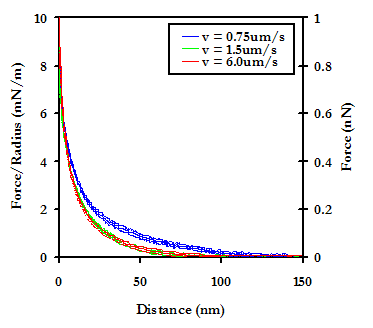
Results on Retract
No effect was observed on retract from varying either the tip compliance or the scan rate.
IV.C.5 Conclusions
The above results suggest that of electrostatic, steric, van der Waals, and hydrophobic interactions, only hydrophobic interactions play a significant role in the intermolecular interaction potential between HSA and PEO. Still, the hydrophobic force is not sufficient to explain either the magnitude or range of the observed repulsive force. The results also suggest that both hydrogen bonding and hydrophobic interactions are key in the nonspecific attractive interactions that form when HSA and PEO come in contact. Formation of these bonds is likely a time-dependent process, although the precise nature of this process has yet to be determined. Finally, within the range of available "soft" cantilevers (i.e ~0.01-0.1 N/m), no effect is observed on the force curve behavior as a function of tip compliance.
Our conclusions thus far point to a set of characteristics we believe are essential for a biomaterial surface to be a successful candidate: 1) The material must be able to complex water molecules extremely well, such that not only would predominantly hydrophobic proteins be repelled by the material, but disruption of the water binding by an incoming protein would pose a significant enthalpic barrier; 2) The material must be flexible and pose a significant barrier via configurational entropy; and 3) The material would ideally be neutral, as all proteins are amphoteric structures, and thus elimination of charge would aid in minimizing attractive interactions. Naturally the next step in this process is to choose possible candidates for superior biomaterial surface coatings, based on the above criteria. The following section details our efforts to do so.
V. Intermolecular Interactions Between HSA and Oligomannose
Derivatives
In
seeking a superior biocompatibile material, we looked first to nature: what
could we learn from biological surfaces that naturally resist protein adsorption?
The glycocalyx, which coats all living cells, performs many functions, including:
keeping cells hydrated, preventing cellular aggregation, and preventing nonspecific
interactions such as protein adsorption. The glycocalyx is dynamic, amphiphilic,
and amphoteric (but possessing a net negative charge), and is heavily populated
by glycoproteins and oligosaccharides. It is believed that the higher oligosaccharides
are particularly effective at preventing nonspecific adsorption and aggregation.
In addition, oligosaccharides fit all of our criteria as outlined in section
IV.C.5. For these reasons, we have begun a
collaboration with Peter Seeberger of the ETH (Zurich, Switzerland), and Daniel
Ratner of the Department of Chemistry at MIT, studying the interactions between
HSA and derivatives of Oligomannose-9 (Man-9), which were custom synthesized
by Ratner, et al.[18] using an elegant novel technique.
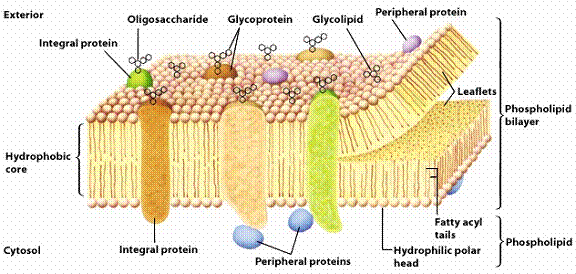
Schematic of the glycocalyx, which coats all living cells (adapted from [19])
Man-9 is present on numerous surfaces throughour the human body, including on the viral envelope of HIV. Located on HIV glycoprotein 120 (gp 120), Man-9 has been identified as the ligand which binds to the CD4 receptor on host cells, allowing the virus to enter host cells and invade the body. A schematic of the function of gp 120 and the structure of Man-9 is shown below.
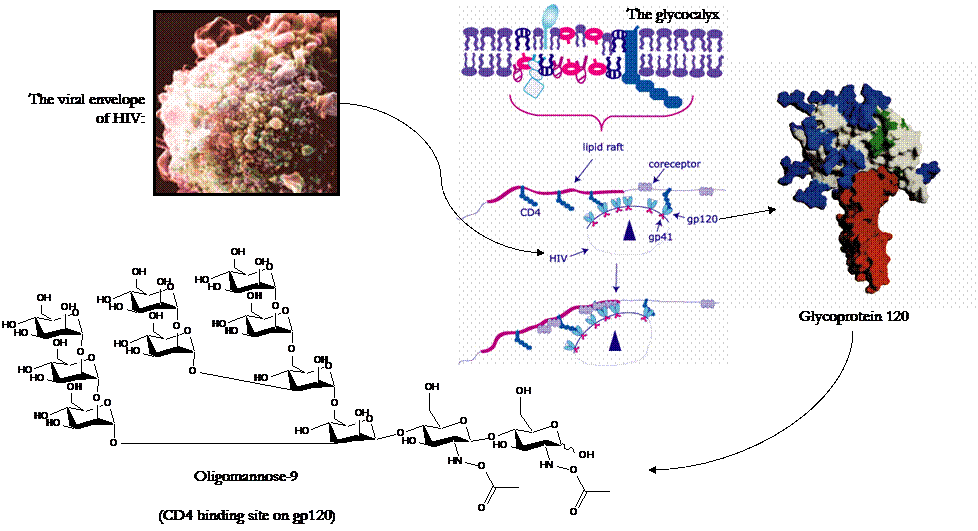
Top Left: Depiction of the viral envelope of HIV (Adapted from [20]); Right: Schematic of the binding of HIV gp210 to the CD4 recptor of host cells (Adapted from [21]; Bottom Left: Structure of Oligomannose-9, the gp120 ligand for the CD4, CCP5, and CXCR4 receptors
We obtained a series of derivatives of Man-9, all of which are present in varying concentrations throughout the human body, and on HIV. Indeed, it is thought that a key factor that makes HIV so "successful" at evading attack by the immune system, is that it mimicks our own biological stealth mechanisms, such as using oligosaccharides to prevent blood protein adsorption! The full structure of Man-9 is present in the highest concentration of all of the derivatives found in the human body, suggesting that the higher oligosaccharides were likely evolutionarily selected because of superior anti-adhesive properties over their simpler counterparts. The derivatives that we obtained were, as mentioned previously, custom synthesized [18], and as a result are lacking the chitobiose tail (the dimer at the right end of the structure above) that links it to cellular and viral surfaces. Instead, our derivatives were functionalized [22] with an n-mercapto tri(ethylene glycol) (EG3: SH-(CH2CH2-O-)-) linker that enables us to attach the derivatives to Au surfaces as was done previously with the alkanethiol SAMs. We ran experiments in PBS (IS = 0.01M, pH = 7.4), and used unmodified Au and an EG3 SAM as controls. EG3 is often used a control in such protein adsorption studies, as it represents one of the most protein-resistant surfaces yet employed in biomaterial science. Structures of the derivatives and controls used in our lab are shown below. It is doubtful that these structures formed well-ordered monolayers, due to their bulkiness and asymmetry.
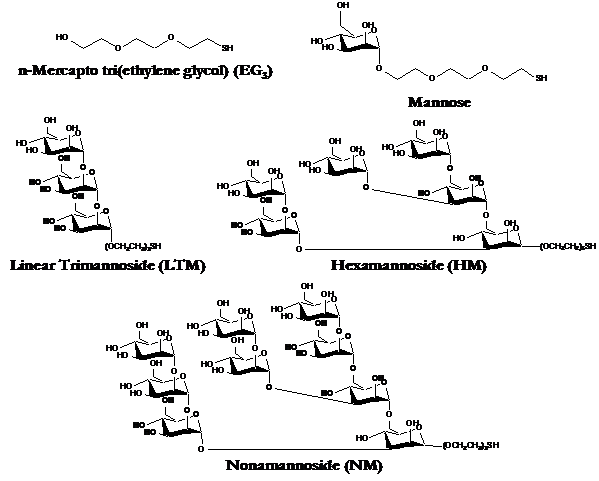
Controls (top) and derivatives of Man-9 employed in our HRFS experiments
Results on Approach
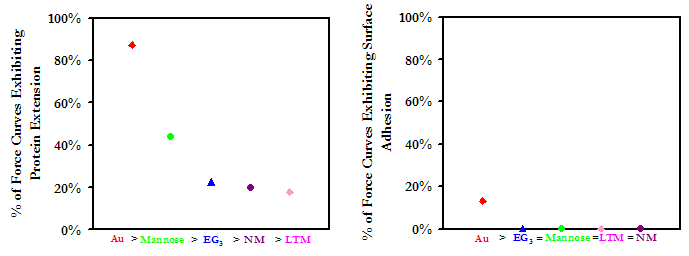
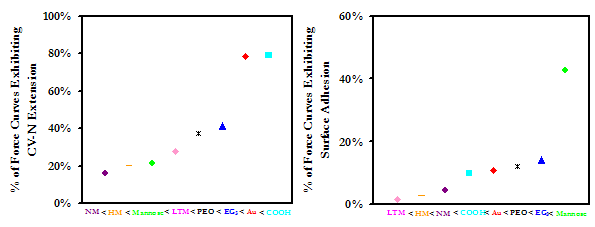
At D > 30nm, all of the oligosaccharide derivatives were more repulsive than either EG3 or Au, with the magnitude of the repulsion increasing with the size of the oligosaccharide. At closer range, D < 20nm, the NM repulsion fell behind EG3, and LTM took the lead. Our results are encouraging, as they suggest that these may possess exceptional protein resistant properties.
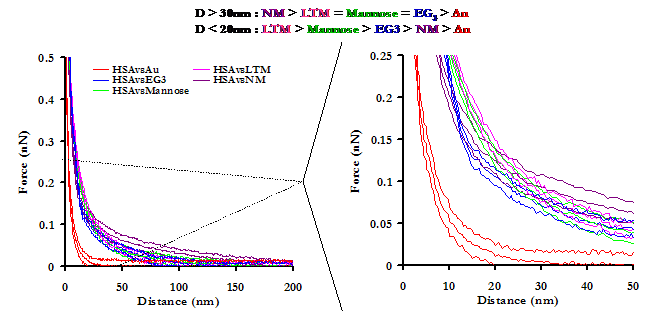
Averaged approach curves of an HSA-grafted probe tip on a variety of surfaces in PBS, with standard deviations. The graph on the right is a magnification of the graph on the left, to aid the reader in interpreting the results
Results on Retract
Once in contact, attractive interactions between HSA and the sugars were formed, enabling protein extension on retract. Compared to the other substrates, NM exhibited the lowest forces and frequencies of adhesion. Our results lead us to suspect that not only do oligosaccharides pose the potential to outperform even the most protein-resistant materials on the market today, but of the candidates tested in this study, the higher oligosaccharides are the most effective at meeting this end. This work is also interesting in the perspective of studying the interaction of blood plasma proteins with HIV, and may provide insights to the stealth mechanisms employed by the virus.
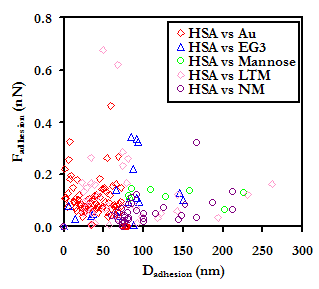
Experimental data points (Above) and frequencies of adhesive events (Below) for an HSA-grafted probe tip on a variety of surfaces, in PBS
V. Intermolecular Interactions Between Oligomannose Derivatives and HIV Therapeutic Drugs
As Man-9 is believed to be directly involved in the binding of HIV to host cells, therapeutic drugs have been developed that target Man-9, binding to it and hence preventing it from binding to CD4. One such drug that shows great promise is Cyanovirin-N (CV-N), a protein originally isolated from extracts of the cyanobacterium Nostoc ellipsosporum, found in blue-green algae. CV-N has been shown to bind irreversibly to gp120 and gp41 on HIV. The protein is made up of a unique sequence of 101 amino acids and forms an elongated tertiary structure with two-fold symmetry, comprised mostly of -sheet structures.[23] The structure, shown below, was adapted from [24].
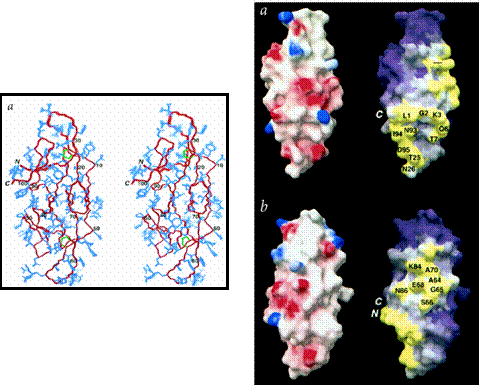
Two views of the three-dimensional structure of cyanovirin-N, a protein found to block HIV interactions with host cells. Left: The backbone is shown in red, the disulphide bridges are shown in green, and all other side chains are in blue; Right: Front and side views of surface maps of cyanovirin-N with respect to a. electrostatic potential, and b. hydrophobic residue clustering. In a., the red regions are negatively charged and the blue regions are positively charged. White regions are neutral. In b., the regions of highest hydrophobicity are colored yellow, and the regions of lowest hydrophobicity are colored purple. White regions are intermediate. (Both images adapted from [24])
The technique of HRFS may easily be extended to the study of specific ligand-receptor interactions, and may provide clues to the binding mechanisms of CV-N and Man-9. We repeated our experiments on the oligosaccharide surfaces in PBS, using a CV-N grafted probe tip. CV-N was obtained through a collaboration with Barry O'Keefe and Kirk Gustafson at the National Cancer Institute in Frederick, MD. Several controls were included in this study, including the R-COO- SAM, the PEO-grafted substrate, the EG3 SAM, and unmodified Au.
Results on Approach:
With the exception of LTM, all of the oligosaccharides were less repulsive than EG3 at D > 25nm, with the magnitude of the repulsion decreasing as the number of saccharides was increased from one to nine. Only Au and R-COO- were less repulsive than NM. These results make sense, in that the affinity of CV-N for each of the structures increases in the order Mannose<HM<NM. It is surprising, however, that LTM exhibited such a strong, long-range repulsion, particularly since LTM has been observed to have the highest affinity for CV-N of all of the structures presented here.[25] At 10nm > D > 25nm, the general trend continues, but Mannose and HM switch order. At D < 10nm, Nearly all the force curves overlap, but NM, Au, and R-COO- exhibit a distinctly lower repulsive force than the other substrates; NM is the least repulsive of the set.

Averaged approach curves of a CV-N-grafted probe tip on a variety of surfaces in PBS, with standard deviations. The graph on the right is a magnification of the graph on the left, to aid the reader in interpreting the results
Results on Retract
Experimental data points of the points of maximum adhesion for each of the substrates is shown below.
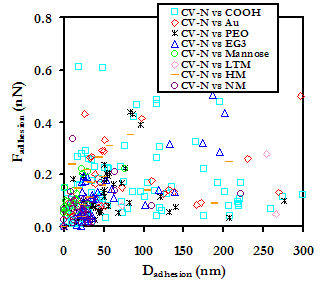
Experimental data points for a CV-N grafted probe tip on various Man-9 derivative substrates, with controls, in BPS
The Au and R-COO- substrates adhere CV-N with the greatest affinity, and show a greater tendency toward protein extension than surface adhesion. All oligosaccharide-modified substrates were less effective at tethering and stretching CV-N than the other substrates, although this may be a result of the unpredictable orientation of CV-N on the probe tip; if the CV-N binding site is only exposed on some of the bound proteins, it may be less probable that every time the probe tip approaches the surface the proteins will be in the proper orientation for binding. In addition, it is quite possible that what is interpreted as surface adhesion on the oligosaccharide susbtrates is in fact ligand-receptor binding that does not result in protein extension. Although Au was able to tether and extend CV-N with high frequency, the strength of the tethering bonds is small compared to other forces of adhesion in the data set. NM was observed to have to largest force of adhesion of all of the substrates. It is also interesting that although the strength of the nonspecific bonding of CV-N to the oligosaccharides is generally mid-to-low range (except NM), the oligosaccharides do exhibit the longest distances of adhesion. These results are currently being examined by us and our collaborators at MIT, the ETH, and the NCI.
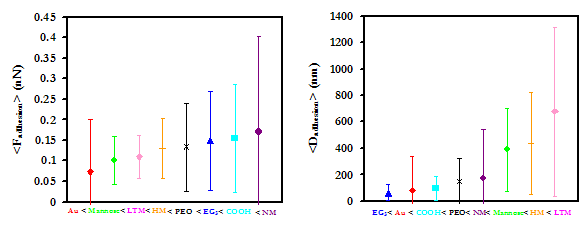
Average forces and distances of adhesion with standard deviations (Above), and the frequencies of adhesion modes (Below) for a CV-N grafted probe tip on Man-9 derivatives and controls
Publications
Rixman, M. A.; Dean, D.; Macias, C. E.; Ortiz, C. Nanoscale Intermolecular Interactions Between Human Serum Albumin and Alkanethiol Self-Assembled Monolayers. Langmuir 2003, 19 (15), 6202-6218. (This paper available in *.pdf here)
Rixman, M. A.; Dean, D.; Ortiz, C. Nanoscale Intermolecular Interactions Between Human Serum Albumin and Low Grafting Density Surfaces of Poly(ethylene oxide). Langmuir 2003, ASAP Article. (This paper available in *.pdf here)
Presentations
Parts of this work have been presented at the following scientific conferences:
April 2001 : 221st American Chemical Society National Meeting, San Diego, CA
June 2001 : Image and Meaning, Massachusetts Institute of Technology, Cambridge, MA
October 2001 : MIT Materials Day, Massachusetts Institute of Technology, Cambridge, MA
October 2001 : Dupont-MIT Alliance Research Symposium, Somerville, MA
March 2002 : Die EuRegionale, Spring-Symposium of the Gesellschaft Deutscher Chemiker JungChemicker Forum, Aachen, Germany
March 2002 : American Physical Society National Meeting, Indianapolis, IA
April 2002 : 4th Annual Northeast Student Chemistry Research Conference, Boston University
August 2002 : Gordon Research Conference : The Science of Adhesion, Tilton, NH
August 2002 : 224th American Chemical Society National Meeting, Boston, MA
November 2002 : Eastern Technical Career Conference, New Jersey
March 2003 : American Physical Society March Meeting, Austin, TX
July 2003 : Gordon Research Conference : Biomaterials - Biocompatibility and Tissue Engineering, Plymouth, NH
REFERENCES:
[1] Curry, S., Mandelkow, H., Brick, P., Franks, N. Nat. Struct. Biol. 1998, 5, 827.; (http://www.rcsb.org/pdb/)
[2] Anderson, J. M. Kottke-Merchant, K. CRC Critical Reviews Biocompatibility 1985, 1, 111-204.
[3] Leckband, D. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 1-26.
[4] Halperin, A. Leckband, D. C. R. Acad. Sci. Paris: Polymers at Interfaces 2000, Series IV, 1171-1178.
[5] Vansteenkiste, S. O.; Corneillie, S. I.; Schacht, E. H.; Chen, X.; Davies, M. C.; Moens, M. Van Vaeck, L. Langmuir 2000, 16, 3330.
[6] Haynes, C. A.; Norde, W. Colloids. Surf. B. : Biointerfaces 1994, 2, 517.
[7] Peters, T. Adv. Protein Chem. 1985, 37, 161.
[8] Soderquist, M. E. Walton, A. G. J. Colloid. Interf. Sci. 1980, 75, 386.
[9] Rixman, M. A.; Dean, D.; Macias, C. E.; Ortiz, C. Langmuir 2003, 19 (15), 6202-6218. (This paper available in *.pdf here)
[10] Seog, J. Dean, D.; Grodzinsky, A.; Plaas, A.; Wong-Palms, S.; Ortiz, C. Macromolecules 2002, 35 (14), 5601-5615. (This paper available in *.pdf format here)
[11] Dean, D.; Seog, J.; Ortiz, C. Grodzinsky, A.; Langmuir 2003, 19 (13), 5526-5539. (This paper available in *.pdf format here)
[12] Ninham, M.; Parsegian, A. J. Theor. Biol. 1971, 31, 405-428.
[13] Christenson, H. K.; Claesson, P. M.; Berg, J.; Herder, P.C.; J. Phys. Chem. 1989, 93, 1472.
[14] Feldman, K.; Haehner, G.; Spenser, N. D.; Harder, P.; Grunze, M. J. Am. Chem. Soc. 1999, 121, 10134-10141.
[15] Oesterhelt, F.; Rief, M. Gaub, H. E. New J. Physics 1999, 1, 6.1-6.11.
[16] Jiang, X. Doctoral Thesis; Department of Chemical Engineering, Massachusetts Institute of Technology: 2002.
[17] Peters, T. All About Albumin: Biochemistry, Genetics, and Medical Applications; Academic Press: 1992.
[18] Ratner, D. M.; Plant, O. J.; Seeberger, P. H. Eur. J. Org. Chem. 2002, 5, 826-833.
[19] Bertozzi Research Group Homepage. “Biomaterial and Interfacial Design”, accessed March 13, 2002: http://www.cchem.berkeley.edu/~crbgrp/research5.htm.
[20] Geriatric Video Productions homepage, ac. E. New J. Physics 1999, 1, 6.1-6.11.
[16] Jiang, X. Doctoral Thesis; Department of Chemical Engineering, Massachusetts Institute of Technology: 2002.
[17] Peters, T. All About Albumin: Biochemistry, Genetics, and Medical Applications; Academic Press: 1992.
[18] Ratner, D. M.; Plant, O. J.; Seeberger, P. H. Eur. J. Org. Chem. 2002, 5, 826-833.
[19] Bertozzi Research Group Homepage. “Biomaterial and Interfacial Design”, accessed March 13, 2002: http://www.cchem.berkeley.edu/~crbgrp/research5.htm.
[20] Geriatric Video Productions homepage, accessed October 4, 2002: http://www.geriatricvideo.com/training/ct.html.
[21] NIH AIDS Reasearch and Reference Reagent Program Homepage, accessed October 4, 2002: http://www.aidsreagent.org/techlib/default.cfm?Action=HIVGraphics
[22] Adams, E.; Ratner, D. M.; Seeberger, P. H.. Manuscript in preparation.
[23] A. J. Bolmstedt, B. R. O'Keefe, S. R. Shenoy, J. B. McMahon, M. R. Boyd, Mol. Pharmacol. 2004, 59, 949-954.
[24] Bewley, C. A.; Gustafson, K. R., Boyd, M. R., Covell, D. G., Bax, A., Clore, G. M. and Gronenborn, A. M. Nat. Struct. Biol. 1998, 5, 571-578.
[25] Ratner, et al. Department of Chemistry, MIT: Unpublished experiments.
Contact Information
Researchers:
Monica A. Rixman, 5th year doctoral candidate in the Program in Polymer Science & Technology, Department of Materials Science and Engineering, MIT : mrixman@mit.edu, (617)258-5934, MIT Room #12-022, 77 Massachusetts Ave., Cambridge, MA 02139. CV available here. Targeted graduation date: February 2004.

Celia Edith Macias, Senior, Department of Materials Science and Engineering, MIT : mec2004@mit.edu, (617)258-5934, MIT Room #12-022, 77 Massachusetts Ave., Cambridge, MA 02139. CV available here.

Mathematical Modeling:
Delphine Dean, 3rd year graduate student in the Department of Electrical Engineering and Computer Science, MIT: finou@mit.edu, (617)253-8385, MIT Room NE47-382, 77 Massachusetts Ave., Cambridge, MA 02139

Faculty Supervisor:
Christine Ortiz, Assistant Professor, Department of Materials Science and Engineering, MIT : cortiz@mit.edu, (617)452-3084, MIT Room #13-4022, 77 Massachusetts Ave., Cambridge, MA 02139

This web page written and produced by Monica A. Rixman, MIT
Last updated: August 4, 2003
 |
 |