HST.175
09/12/00
1:30 to 2.30
Shiv Pillai
ANTIBODIES
(Recommended reading: Abbas et al., Chapter 4; Janeway and Travers, Chapter 3)
The session on antibodies will be followed by a discussion of some aspects of cell biology. Understanding antibody synthesis and function can help provide a broad picture of how secretory and membrane proteins are transported within a cell and how signals are transmitted from the cell surface to the nucleus.
Antibodies protect us from a vast variety of pathogens. Indeed the antibody repertoire is immense - the binding or combining sites of antibodies may be able to recognize somewhere in the range of 10 million different shapes. Antibodies share with agglutinins all the features that contribute to the elimination of pathogens, and can also contribute to host defense in some ways that the innate immune system cannot.
Antibodies are also called immunoglobulins. A very crude electrophoretic fractionation (separation on the basis of charge) of serum proteins separates albumin from other serum proteins collectively called globulins. The globulins are also further characterized on the basis of charge as a, b and g -globulins, g-globulins being the most positively charged. This latter fraction was discovered to be made up largely of antibody molecules. The existence of antibody secreting tumors known an myelomas or plasmacytomas greatly facilitated the study of immunoglobulins and their structure. Tumors are in general derived from a single cell. A single cell and its progeny are generally referred to as a clone and tumors are therefore clonal (or monoclonal) outgrowths. Myelomas and plasmacytomas are derived from differentiated antibody secreting B lymphocytes known as plasma cells These plasma cell tumors each produce large amounts of a single or monoclonal antibody.
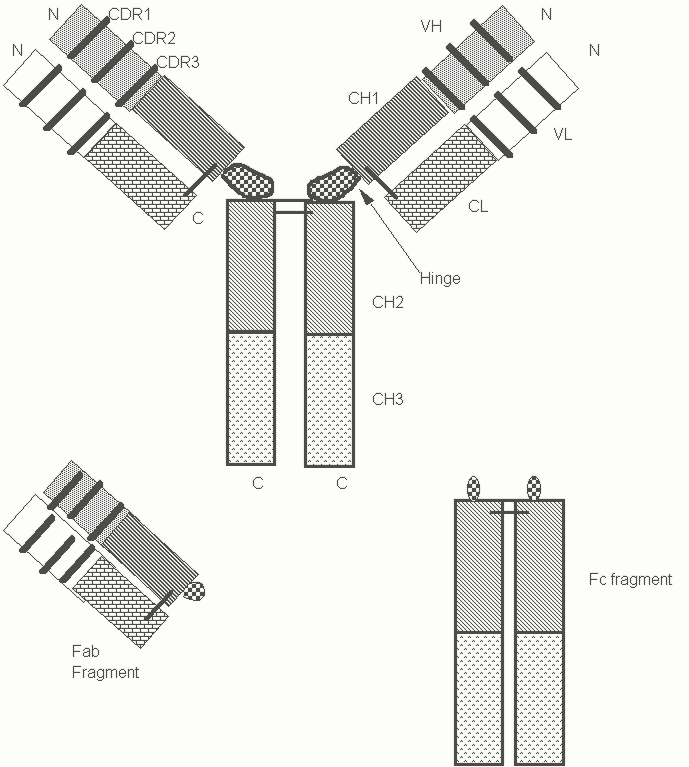
From our knowledge of the biology of agglutinins that mediate innate immunity, we might well make certain predictions about antibody structure and function. Antibody molecules secreted by individual B lymphocytes would, of course, be predicted to differ in their combining sites for antigen. It might perhaps be logically assumed that all antibody share common features that enable them to agglutinate particulate antigens, to assist in the process of phagocytosis (by opsonization) and to bind to complement proteins. Indeed, antibodies possess all these predicted features. The basic structural unit of all antibodies is, as shown in Figure 1, a tetramer of two heavy and light chains, containing two identical binding sites for antigen. This bivalent structural unit is Y shaped with the two binding sites set sufficiently apart to facilitate agglutination. The portions of the heavy chain and light chain that are involved in antigen recognition are at the N-terminal ends and are referred to as V (or variable) domains. The remaining portions of the heavy and light chains are referred to as constant (or C) regions. Each light chain constant region contains a single constant domain (CL). A domain refers to a portion of a protein which can be separated from the rest of the molecule and still fold into its correct shape. The domains in immunoglobulin molecules have a typical three dimensional structure which is now referred to as an immunoglobulin domain and is found in a variety of proteins (many of which existed before the evolution of immunoglobulins). Each heavy chain constant region is made up of three or four domains (CH). The portion of each heavy chain that is associated with the light chain is the VH domain and the CH1 domain. A cysteine residue in the CH1 domain forms a covalent disulfide bridge with a cysteine residue towards the C-terminal end of the CL domain. When one class of immunoglobulin molecules known as IgG molecules are cleaved with a proteolytic enzyme called papain, these antibodies are cleaved into two identical fragments known as Fab (Fraction antigen binding) fragments and a single Fc (Fraction crystallizable) fragment. Cleavage with pepsin yields an F(ab)2 fragment and an Fc fragment. Each Fab fragment corresponds to an arm of the antibody Y and is made up of a light chain covalently associated (via a disulfide bridge) with a portion of the heavy chain containing the VH and CH1 domains. The Fc portion corresponds to the stem of the Y and is a dimer of the CH2 and CH3 domains of IgG covalently united via disulfide bridges between the two heavy chains.
The variable domains of both heavy and light chains contain a large number of residues that are identical or highly conserved between different antibodies. These are referred to as framework regions. The variable nature of these domains is contributed to by stretches of amino acids that differ from one antibody variable domain to another. These residues make up three hypervariable regions or complementarity determining regions or CDRs. In both the heavy chain and the light chain CDR3 is the most variable of the hypervariable regions. Every constant or variable region domain forms an immunoglobulin fold which looks a lot like two very similar slabs slapped together at a slight angle. Each slab is made up of strands of a polypeptide strung up and down in succession to make a structure known as a b-sheet. The immunoglobulin fold itself is made up of these two slabs of b-sheets compressed against each other (known as an antiparallel b-barrel). At the top and bottom of this sandwich are loops that maintain continuity between individual strands of these b-sheets. When one examines the structure of a variable domain the three CDRs form loops of highly variable configuration that protrude from the "top" of a very conserved immunoglobulin fold structure. It is these very variable CDR loops of a VH and a VL domain that combine to form a binding site for antigen.

Figure 1: Schematic view of the structure of an IgG antibody.
If an antibody is directed against a small antigenic determinant or epitope the binding site formed by the three CDR loops may resemble a relatively tight pocket into which the epitope might fit. If the epitope is somewhat larger such as the rough terrain forming an antigenic "patch" on the surface of a protein, the combining site of the antibody may be formed by an outward splaying of the six CDR loops. The six hypervariable regions from the heavy and light chains combine to form a complementary surface to that of the protein antigenic determinant with some of the little hills on one surface dipping into the valleys of the other and vice versa
Antibodies exist as a number of classes or isotypes (defined on the basis of their heavy chains). The antibody isotype that is made early in an immune response is of the IgM class. Later in an immune response a given B lymphocyte may "switch" from producing IgM antibodies against a specific antigen, to producing antibodies of other isotypes, such as IgG, IgA or IgE, also directed against the original antigen. Class switching of antibodies typically occurs in responses to protein antigens that are driven in part by signals from T lymphocytes. Another important phenomenon that tends to occur in immune responses driven by T cell derived signals is a process by which mutations are introduced into the genes encoding the variable portion of the antibody heavy and light chains. This process is referred to as somatic mutation. As we will discuss in later lectures, the selection by antigen of B lymphocytes whose mutated receptors "fit" the antigen most tightly results in the further differentiation of only those B cells which make antibodies of high affinity toward their cognate epitopes. This process is also described as affinity maturation. Since class switching occurs in cells that also undergo affinity maturation, IgG, IgE and IgA antibodies are likely to have combining sites which fit the antigenic determinant more tightly than the original IgM antibody made by the same B cell before it switched and mutated its immunoglobulin genes.
Although IgM antibodies tend to have individual binding sites that have a lower affinity for antigen than the corresponding combining sites on IgG antibodies the valency of secreted IgM antibodies permits them to bind to antigens quite well. IgG antibodies are each made up of two identical heavy chain molecules and two identical light chain molecules. When purified by being sedimented at very high speeds in an ultracentrifuge, IgG molecules are described as being 7S antibodies. S represents a Svedberg unit, named after the Swedish scientist who pioneered the use of the ultracentrifuge and is a measure of the hydrodynamic properties of a protein which generally reflect the size of the protein being examined. Secreted IgM antibodies are generally referred to as 19S immunoglobulins. They are made up of pentamers and hexamers of the basic 7S unit. Pentameric IgM therefore contains ten combining sites and is able to hold on to an antigen more tightly because of this multiplicity of sites, even though individual sites are of relatively low affinity. It is this overall ability to bind antigen which is measured as avidity.
The structural features of each antibody class is determined by a distinct type of heavy chain protein. The VH domain of a given antibody is originally physically in continuity with CH domains of the m heavy chain. Antibodies of each of the other classes and sub-classes have distinct heavy chain proteins. Although a given B cell starts out making IgM and may later secrete IgG, IgA or IgE it always makes either a k or a l light chain. These two types of light chain are referred to as light chain classes or isotypes. There are no major functional differences that can be attributed to k or l light chains.
A brief note on gene expression and protein structure:
The primary structure of a protein is its amino acid sequence. The most common way a protein's primary structure is determined today is by cloning a cDNA that encodes this protein. It is useful to have a clear picture of what is meant by the terms genomic DNA and cDNA. A promoter is a site at which (DNA dependent) RNA polymerase initiates transcription making an RNA copy of one strand of genomic DNA. Most eukaryotic promoters contain an AT rich sequence known commonly as a TATA box. Proteins that contribute to the basal transcription machinery direct RNA polymerase II (the enzyme responsible for initiating transcription of most genes) to initiate transcription about 30 basepairs downstream of the TATA box at the transcriptional initiation site (cap site). The primary transcript is processed in three ways - it is capped at the 5' end, spliced (introns are removed) and polyadenylated at the 3' end. The processed transcript is known as mRNA (messenger RNA). In the laboratory an enzyme known as reverse transcriptase (RNA dependent DNA polymerase) is used to convert mRNA into a complementary copy known as a cDNA. The set of cDNAs from B lymphocytes, for instance will include antibody genes but these genes will not be found in cDNA libraries from hepatocytes or muscle for instance. Looking at a cDNA sequence (or the predicted primary structure) can often reveal important information about the protein of interest. Homology to known cDNA sequences can yield clues to structure and function.
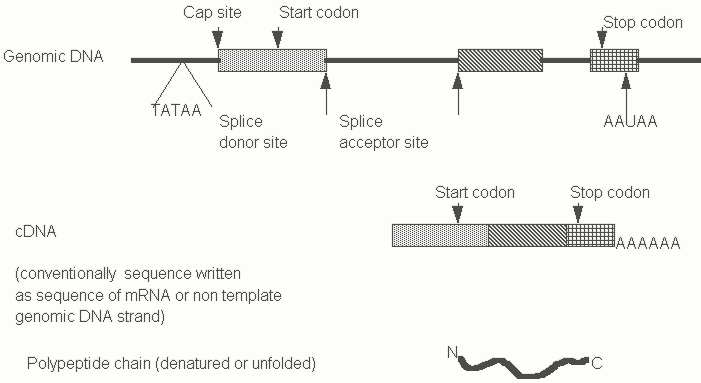
Figure 2. A very basic view of a genomic DNA template
.
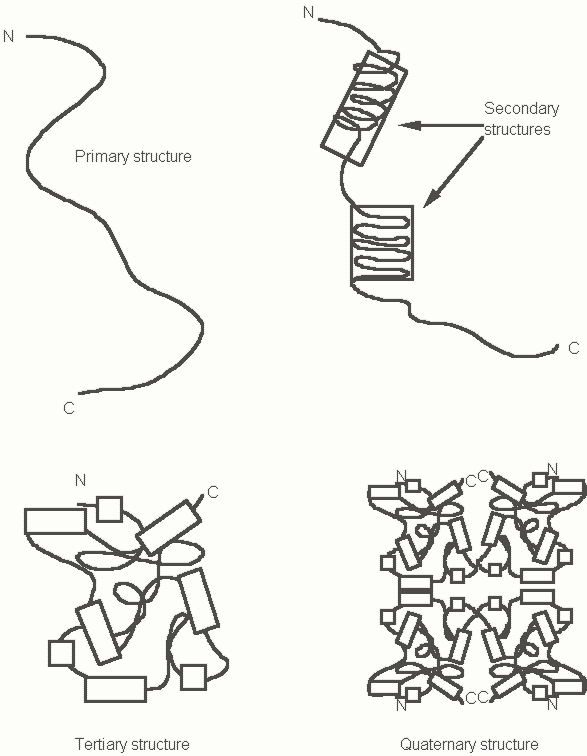
Figure 3. A review of the fundamentals of protein structure
The nature of the side chains or R groups of the amino acids that make up a polypeptide chain determine the folding pattern of a given protein. Certain amino acids can contribute to the formation of local secondary structures which may take the form of an a-helix or a b-sheet. The secondary structures are held together by hydrogen bonds between the NH groups and C=O groups of adjacent amino acids. Alpha helices are right handed polypeptide spirals. Certain amino acids are preferred in alpha helices. Beta sheets are made up of beta strands which run parallel to one another and are connected by intervening loops of amino acids. Regions of secondary structure are packed together in the folded globular tertiary structure of a protein. Proteins that are made up of more than one subunit are usually arranged in a symmetric manner around a central axis and exhibit quaternary structure. Multi-subunit proteins are sometimes referred to as oligomeric proteins. If all the subunits are identical these proteins are homo-oligomers; if not they are called hetero-oligomers. Subunits may be non-covalently associated or bridged by disulfide bonds between cysteine residues.
Some structural feature of antibodies will be discussed.
Objectives/ Study questions:
Some aspects of molecular and cell biology should be reviewed briefly if necessary.
1. Review basic concepts in gene expression and translation. What is a promoter? Where does transcription initiate from? What constitutes the translational start site ? What is RNA processing?
2. Review some basic concepts in protein structure. What do you mean by primary, secondary, tertiary, and quaternary structure? You should know the basic structure of an amino acid, be able to define what an "R" group is, and be able to identify in a general way some of the characteristics of R groups - e.g. lysine and arginine have positively charged R groups.
3. What is the secretory pathway? What is a signal or a leader peptide?
4. Outline the domain structure of an IgG antibody.
5. What is the difference between the structure of IgG, IgM, and IgA?
6. How are membrane and secretory antibodies generated?
7. Define idiotypes, allotypes, and isotypes.
8. Look into the functions mediated by the effector domains of antibodies (these will be considered in detail in a later class).