HST 175- Fall 2000
9/14/00
1.30 to 4 p.m.
Shiv Pillai
Antigen Receptors and the Generation of Diversity
(Recommended reading: Abbas et al., 4th edition, Chapters 6 and 7 Janeway et al., 4th edition, Chapters 3 and 4)
The antigen receptors on B and T lymphocytes (the B Cell Receptor or BCR, and the T Cell Receptor or TCR) are broadly similar in structure and are believed to initiate signaling in similar ways. The antigen binding chains of these receptors do not directly contact cytosolic signaling molecules, but interact with accessory proteins which are also anchored in the plasma membrane and whose cytoplasmic tails contain motifs known as ITAMs (Immunoreceptor Tyrosine based Activation Motifs). ITAMs contain the following consensus sequence:
YxxL/Ix6-8YxxL/I
These motifs are found in proteins associated with a number of signaling receptors in the immune system. When tyrosine residues in "YxxL/I" tetrapeptides are phosphorylated, they may be recognized by specific SH2 domains present in certain proteins. Phosphorylated ITAMs are therefore capable of recruiting molecules that contain specific SH2 domains to activated antigen receptors.
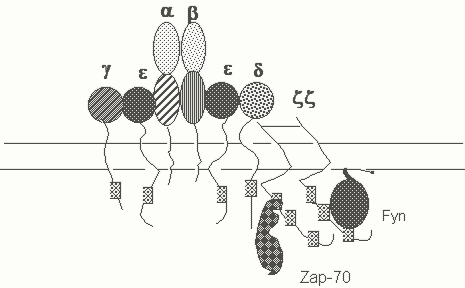
The TCR is made up of either a and b chains (in ab T cells) or g and d chains (in gd T cells). These antigen binding transmembrane polypeptides are linked to one another by a disulfide bridge. Each heterodimer is associated with the CD3 complex made up of integral membrane proteins which contain ITAMs in their cytoplasmic tails (see Figure 1). The pre-T receptor is generated only in ab lineage cells prior to TCR a chain gene rearrangement, and will be discussed in detail in the lecture on T cell development. This receptor consists of the TCR b chain linked to a surrogate a chain known as preTa and the non-covalently associated CD3 complex. The CD3 complex consists of CD3g, CD3d, CD3e, and CD3z chains. z chains generally exist as disulfide linked homodimers and contain three ITAMs in their cytoplasmic tail regions; a small proportion of TCRs may be associated with z-h heterodimers. The h chain is the product of an alternatively spliced version of the z chain gene. The g,d, and e chains of the CD3 complex each contain a single cytoplasmic ITAM.

B lineage pre-antigen receptors (the pro-B receptor and the pre-B receptor) as well as the B cell antigen receptor, share very similar structures. The antigen receptor on B cells is made up of membrane immunoglobulins (IgM and IgD in naive B cells; IgG, IgA, or IgE in some activated and memory B cells) associated with a disulfide linked heterodimer made up of two integral membrane glycoproteins, Iga and Igb. These latter proteins each contain an ITAM in their cytoplasmic tails. The pre-B receptor lacks conventional light chains, but contains pre-B specific surrogate light chains. Pro-B and Pre-B receptors will be discussed in greater detail during the lecture on B cell development. The cytoplasmic tails of the m and d heavy chains contain only three amino acids (KVK) which basically function as a "stop transfer" sequence during the process of translocation into the ER and do not present a significant surface for the association of these receptors with cytosolic signal transducers. The ITAMs in Iga and Igb are critical for signal transduction) by the B cell receptor (Figure 2).
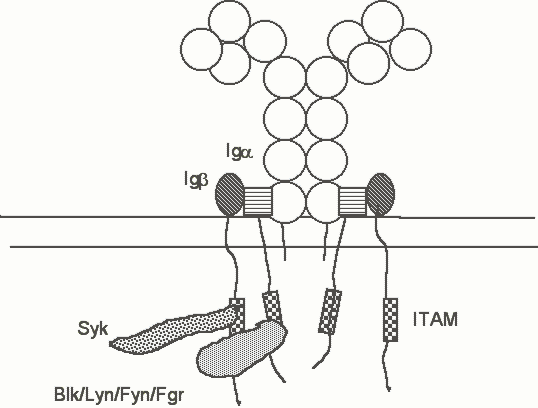
Figure 2. A schematic view of the B cell receptor. Iga and Igb form disulfide linked heterodimers which are non-covalently associated with membrane immunoglobulin H-L tetramers. Blk, Lyn, Fyn, Fgr, and Lck are Src family kinases which associate with the B cell receptor. These kinases are myristylated (not shown) and associate with the inner face of the plasma membrane. Their N-termini are involved in the interaction with the spacer portion of the ITAM. Syk probably associates via its tandem SH2 domains with constitutively phosphorylated tyrosine residues in the ITAMs
Although antigen receptors may potentially initiate signals in multiple ways, the major mechanism by which signals are generated is the activation of tyrosine phosphorylation. Src family kinases associate with the antigen receptor even in quiescent cells. The cytoplasmic tails of coreceptor proteins (discussed below) also bind to Src family kinases. Some activation of antigen receptors occurs even in naive B lymphocytes and might represent "tonic" signaling. In T cells, it is clear that the "tickling" of naïve TCRs is mediated by self-MHC molecules presenting self-peptides. In B cells it remains to be established if signaling in naïve cells is self antigen-driven or "tonic", an issue that will be discussed in Chapter Seven. These low level "tonic" or self-antigen driven signals might contribute to the just detectable but constitutive phosphorylation of tyrosine residues in ITAMs. This phosphorylation probably permits the minimal, but detectable, recruitment to the receptor, in "resting" lymphocytes, of members of the Syk/Zap 70 family of tyrosine kinases.
Src family kinases (Blk, Lyn,Lck, Fyn, Fgr) associate with the inner face of the plasma membrane via N-terminal lipid modifications and are loosely associated with antigen receptors in non-stimulated B and T cells. In the case of the B cell receptor (BCR) this association involves an interaction between the unique N-terminal region of the Src family kinase with a loop region between the tyrosine residues of the ITAM motif in the cytoplasmic tails of Iga and Ig. In a similar fashion, Lck associates with the cytoplasmic tails of CD4 and CD8 co-receptors in T cells. Fyn probably associates with the ITAMs of CD3 chains, particularly z and e, although this interaction might be indirect.
The generation of diversity
The tremendous diversity of the immune system is largely generated in developing B and T cells by the process of gene rearrangement. This process is intrinsically designed to be somewhat inaccurate - the addition or removal of bases during the joining process contributes to the generation of diversity. As a result rearrangement is intimately tied to selection processes. Cells that make productive rearrangements are selected to survive and differentiate further while cells that fail to do so die by default. The rearrangement of immunoglobulin and T cell receptor genes is initially activated in pre-B and pre-T cells and because it involves the joining of V (variable) region genes to D (diversity) segments, and of D segments to J (junctional) regions, this process is known as VDJ recombination. VDJ recombination is actually activated more than once during B cell development. At the pre-B stage VDJ recombination generates rearranged heavy and light chain loci. At the immature B stage, cells that encounter multivalent self-antigen are given an opportunity to reform themselves or to die. Signals delivered by the B cell receptor reactivate the process of VDJ recombination; cells that successfully make a new receptor by a process that is known as "receptor editing" may evade death and become useful citizens.
V(D)J Recombination
The repertoires of B cells and both ab and gd T cells are primarily generated by V(D)J recombination. In the case of antibody genes, the heavy chain locus and the light chain loci are on different chromosomes. In the heavy chain locus a large number of V (variable region) genes are tandemly arranged upstream of about 20-30 D (diversity) segments. A few J segments are separated by the J-C intron from heavy chain constant region (C) exons. Both combinatorial and junctional mechanisms contribute to diversity. D segments are found in the Ig heavy chain locus and the TCRb and d loci. Two VDJ recombinational events are involved in the joining process at these loci. D segments are lacking in the Ig light chain, TCRa and TCR g loci.
Rearrangement is an ordered process. The first rearrangement that occurs in a cell committed to the B lineage, for instance, is a D-J rearrangement at the heavy chain locus. This event is followed by V to DJ rearrangement and sequential rearrangement at the k and l light chain loci.
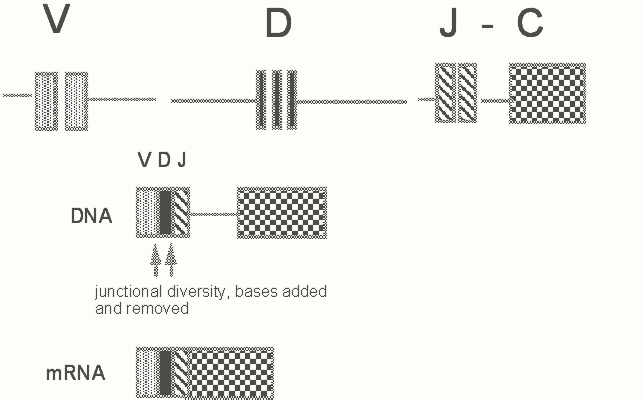
Figure 3. Schematic view of the organization of the Ig heavy chain locus (in man there are about 1000 V genes, 6 functional JH segements and about 30 D segments). Upstream of each V gene is a leader exon which encodes a signal peptide for translocation of the heavy chain protein into the endoplasmic reticulum.
All immunoglobulin and T cell receptor gene segments which undergo rearrangement are flanked by recombination signal sequences (RSSs). Immediately adjacent to the coding region is a conserved heptamer (CACTGTG or its reverse complement CACAGTG) followed by a spacer and then a relatively conserved nonamer (GGTTTTTGT or its reverse complement ACAAAAACC).
The spacer in an RSS is either approximately 12 bp in length or approximately 23 bp in length. RSSs witth 12 bp spacers recombine only with RSSs with 23 bp spacers (the 12/23 spacer rule).
The entire process of VDJ recombination can be divided into three steps:
1. Cleavage of the DNA to generate a double stranded break
2. Processing of the cut ends - primarily to generate greater diversity
3. Joining the processed ends.
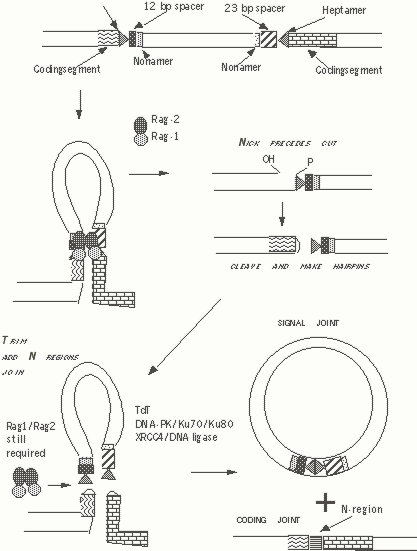
Figure 4. Current view of role of RAG-1 and RAG-2 in VDJ recombination. DNA is cut at the heptamer coding region boundary by RAG-1 and RAG-2. Coding ends form hairpins while signal joints (on right) are formed between flush cut heptamer ends.
However the division of the process into these three steps is somewhat artificial. Molecules such as Rag-1 and Rag-2 which are involved in the cutting step are also probably required for joining.
Cleavage and generation of a double stranded break
The RAG-1 and RAG-2 proteins recognize RSSs and specifically cut the DNA in a flush end between the heptamer and the coding sequences.RAG-1 and RAG-2 may form a dimer and are both needed for recombination. These proteins are nuclear proteins; little is understood about the structural basis for the function of Rag-1 and Rag-2.
It has become clear that RAG-1 and RAG-2 alone are not only sufficient to cleave DNA at heptamer-coding region junctions but that this complex is required for subsequent events during VDJ recombination possibly because it helps to hold the coding junctions and the signal junctionss together in a sort of synaptic complex while other gene products complete the process of VDJ recombination.
RAG-1 binds to nonamer sequences in RSSs and probably recruits RAG-2. It is possible that a tetrameric RAG complex is formed. The RAG-1 /RAG-2 complex may bring together RSSs containing 12 bp and 23 bp spacers and probably only when this has been achieved, initiate the cutting process. At the heptamer-coding sequence junction, RAG-1/RAG-2 first makes a nick and the free nicked end attacks the other strand generating a closed hairpin structure at the coding end. A flush heptamer signal end results from the double-stranded break.
Processing of broken DNA ends
Blunt signal ends are joined without any processing. However the hairpin coding ends are opened up and nucleotides may be added to or removed from these ends resulting in greater immune diversity (an a fair number of out-of frame rearrangements). Very little is understood in molecular terms as to how nucleotide loss occurs. Presumably endo- or exo-nucleases are involved which only recognize coding/hairpin ends. It is possible that nucleotide loss occurs directly as a result of the joining mechanism. Some deletions would be expected when recombination is directed by short sequence homologies.
Template-dependent and template-independent mechanisms exist
for the addition of nucleotides to coding ends. Terminal
deoxynucleotidyl transferase (TdT) is an enzyme responsible for N
region addition. N regions are short stretches of bases, ususally GC
rich, that are found at junctions and which are added in a template
independent manner. Template dependent nucleotide addition occurs
when hairpins are cleaved eccentrically resulting in a few additional
nucleotides in a single stranded extension. These extra nucleotides
are retained in coding joints and are called P nucleotides. Opening
of hairpins may require DNA-PK an enzyme that is required in the
joining process.
Joining of broken DNA ends
Proteins that are involved in Double Strand Break repair (DSB) are critical for the joining process in VDJ recombination. These genes include Ku70, Ku 80, DNA dependent protein kinase ( DNA-PK), XRCC4 and DNA ligase IV. DNA-PK is the scid gene product and forms a complex with Ku70 and Ku80.The scid gene is so named because it is defective in murine Severe Combined Immune Deficiency. In the absence of DNA-PK/scid, Ig and T cell receptor rearrangement is extremely inefficient and scid mice basically lack B and T cells. The DNA-PK/ Ku70/Ku80 complex is known to play a role in DNA repair. DNA-PK is a member of a growing family of dual lipid/protein kinases, prototypic members including PI-3 kinase and the ATM (ataxia telangiectasia mutated) protein. The Ku 70 and Ku 80 proteins are non-catalytic subunits of DNA-PK.
Selected reviews:
Gellert, M. (1996). A new view of V(D)J recombination. Genes Cells 1, 269-275.
Gellert, M. (1992). V(D)J recombination gets a break. Trends Genet. 8, 408-412.
Jeggo, P. A. (1997). DNA-PK: at the crossroad of biochemistry and genetics. Mutation Res. 384, 1-14.
Oettinger, M. A. (1996). Cutting apart V(D)J recombination. Curr. Opin. Genet. 6, 141-145.
Schatz, D. G. (1997). V(D)J receombination moves in vitro. Semin. Immunol. 9, 149-159.
Sleckman, B. P., Gorman, J. R., and Alt, F. W. (1996). Accessibility control of antigen-receptor variable region gene assembly: role of cis-acting elements. Annu. Rev. Immunol. 14, 459-481.
Tonegawa, S. (1983). Somatic generation of antibody diversity. Nature 302, 575-581.
Weaver, D. T. (1995). What to do at an end: DNA double strand break repair. Trends Genet. 11, 388-392.
Objectives/ StudyQuestions
1. What function is mediated by an ITAM motif?
2. What are coreceptors? How are they different from costimulatory receptors?
3. What are tyrosine kinases? What is the temporal order in which different tyrosine kinase families are activated downstream of antigen receptors?
4. What is the 12/23 rule?
5. Name the lymphoid-specific factors involved in V(D)J recombination. What do they do?
6. What are N regions? What type of diversity do they contribute to?
7. What is the order of rearrangement of Ig genes and gene segments in the B lineage?
8. Which chain of the T cell receptor is "developmentally homologous" to the Ig heavy chain?
9. What ubiquitous proteins are required to complete the process of V(D)J recombination?