Evolutionary Mean Field Dynamics
![]() Assume a fitness landscape for viral sequence (single protein or whole genome),
Assume a fitness landscape for viral sequence (single protein or whole genome),
[binary (0 for consensus, 1 for mutant) of length L] including one site and two site terms.
Add an additonal cost from epitopes targeted by the host immune system (mostly T cells for HIV):
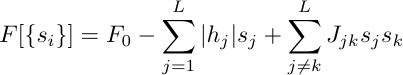
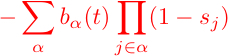
Use the
equations to compute the time evolution of single site mutation probabilities
![]() ,
,
where the `mean-field approximation' from statistical physics gives
=h_i.jpeg)
_pro.jpeg)
The essence of the `mean-field approximation' is the neglect of the correlations betweem sites, setting
![]()
Such `linkage equilibrium' is achieved in population genetics if the sequence elements are completely reshuffled post replication, i.e. for infinite recombination rate [Neher & Shraiman, Rev. Mod. Phys. 83, 1283 (2011)] (offline)
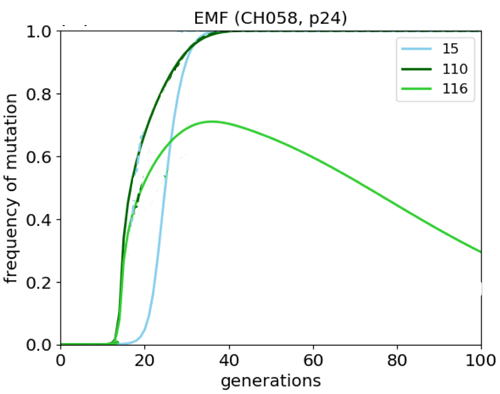
![]() How well does this approach work in an actual patient?
How well does this approach work in an actual patient?
Patient CH58 [Liu et al, J. Clin Invest 123, 380 (2013)] infected with HIV
Blood samples extracted at several time points to determine HIV sequences; T cell responses were also mapped.

Putative fitness landscape of p24 was constructed using maximum entropy approach on sequence diversity.
Given the many approximations involved, the results of mean-field dynamics are encouraging:
 vs.
vs.