 Figure 1: Escher's
vision of topological aspects of DNA.
Figure 1: Escher's
vision of topological aspects of DNA.
 Figure 1: Escher's
vision of topological aspects of DNA.
Figure 1: Escher's
vision of topological aspects of DNA.
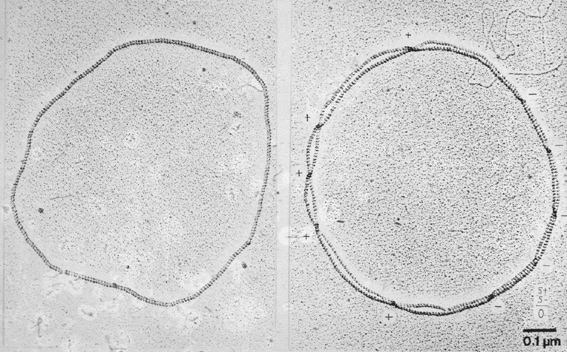 |
Figure 2: Coating of DNA with RecA allows us to distinguish over- and under-crossings. |
| Figure 3: Topoisomerase I induces formation of left- and right-handed DNA trefoils with equal probability when acting on nicked circular double-stranded DNA molecules. | 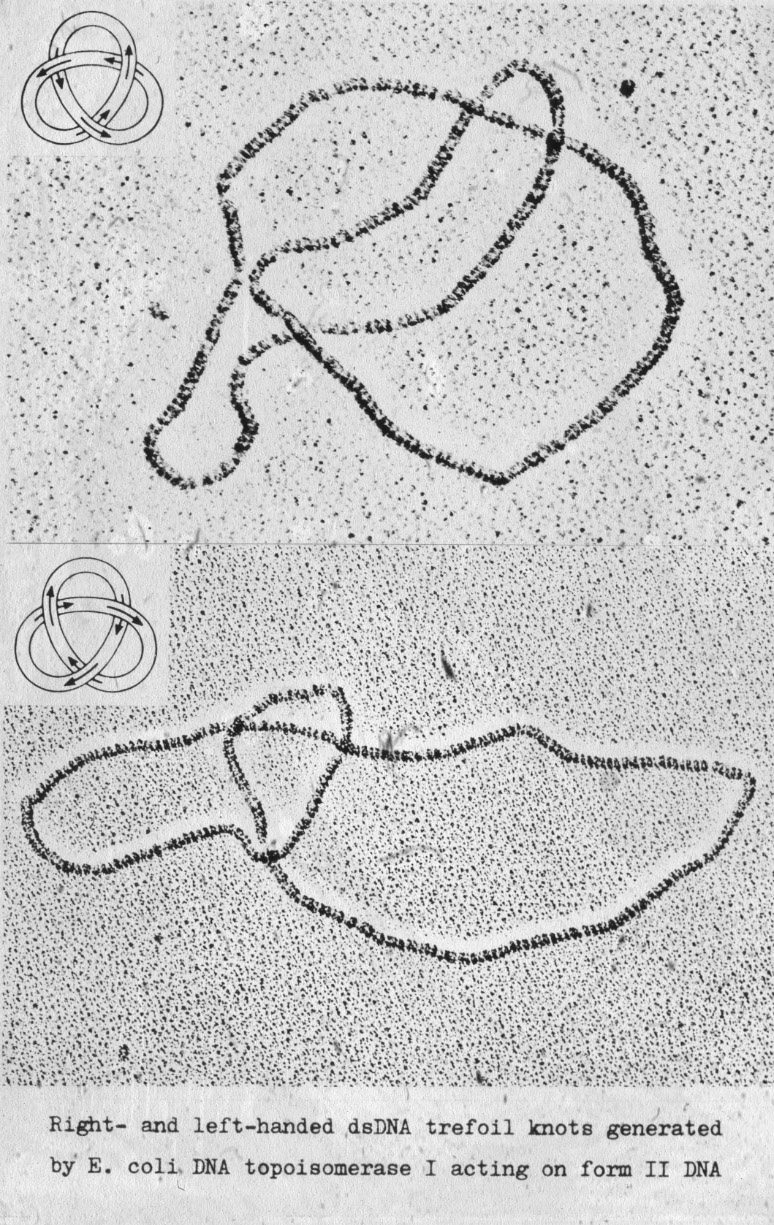 |
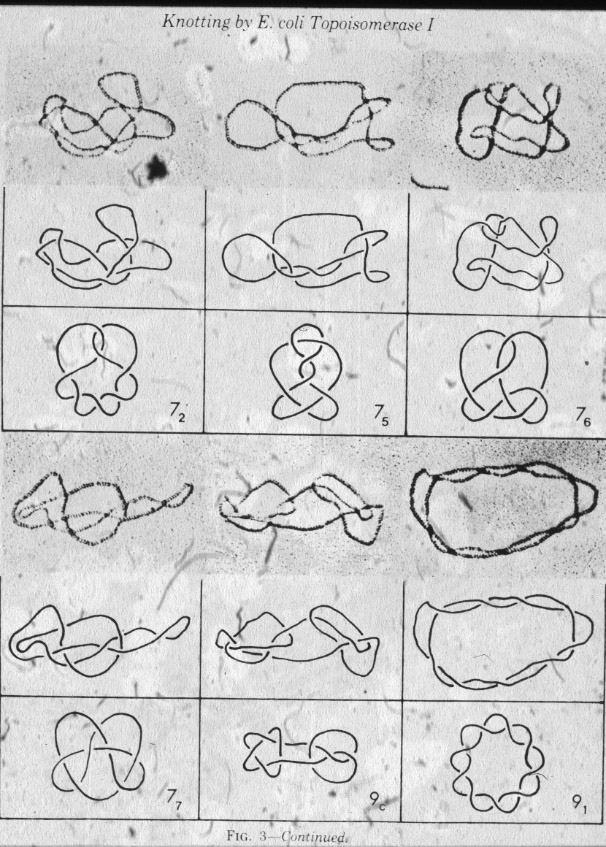 |
Figure 4: Topoisomerase I produces all possible knots when acting on nicked circular double-stranded DNA molecules. |
| Figure 5: Schematic drawing of the site specific intermolecular recombination reaction leading to the integration of one circular DNA molecule into another one. | 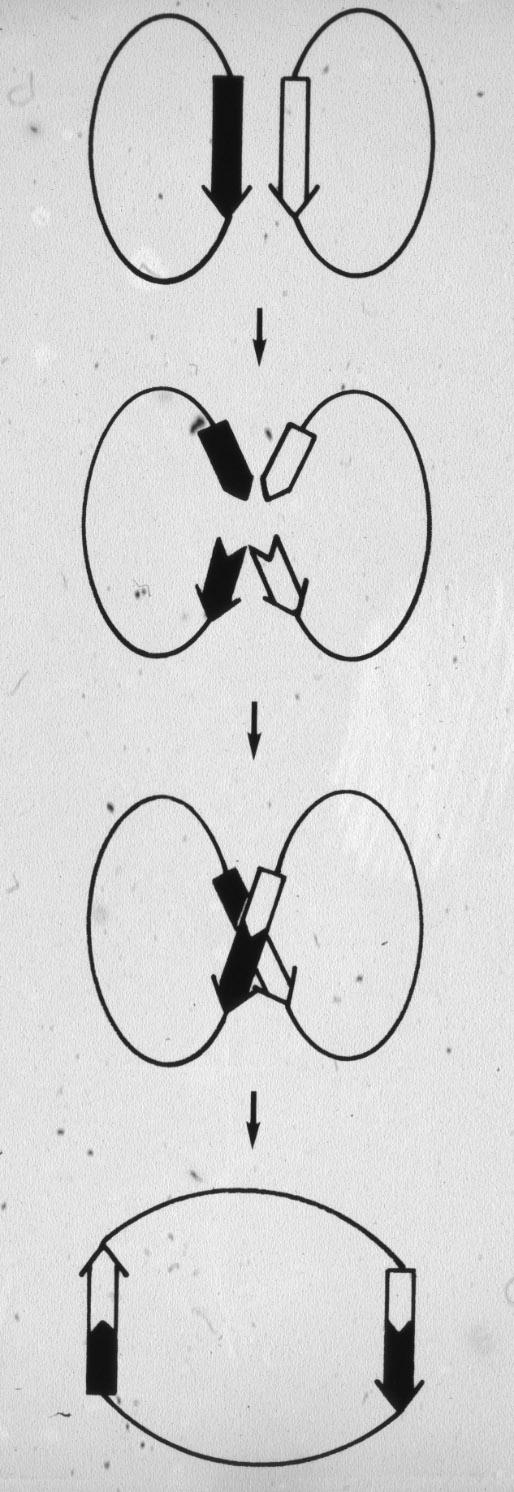 |
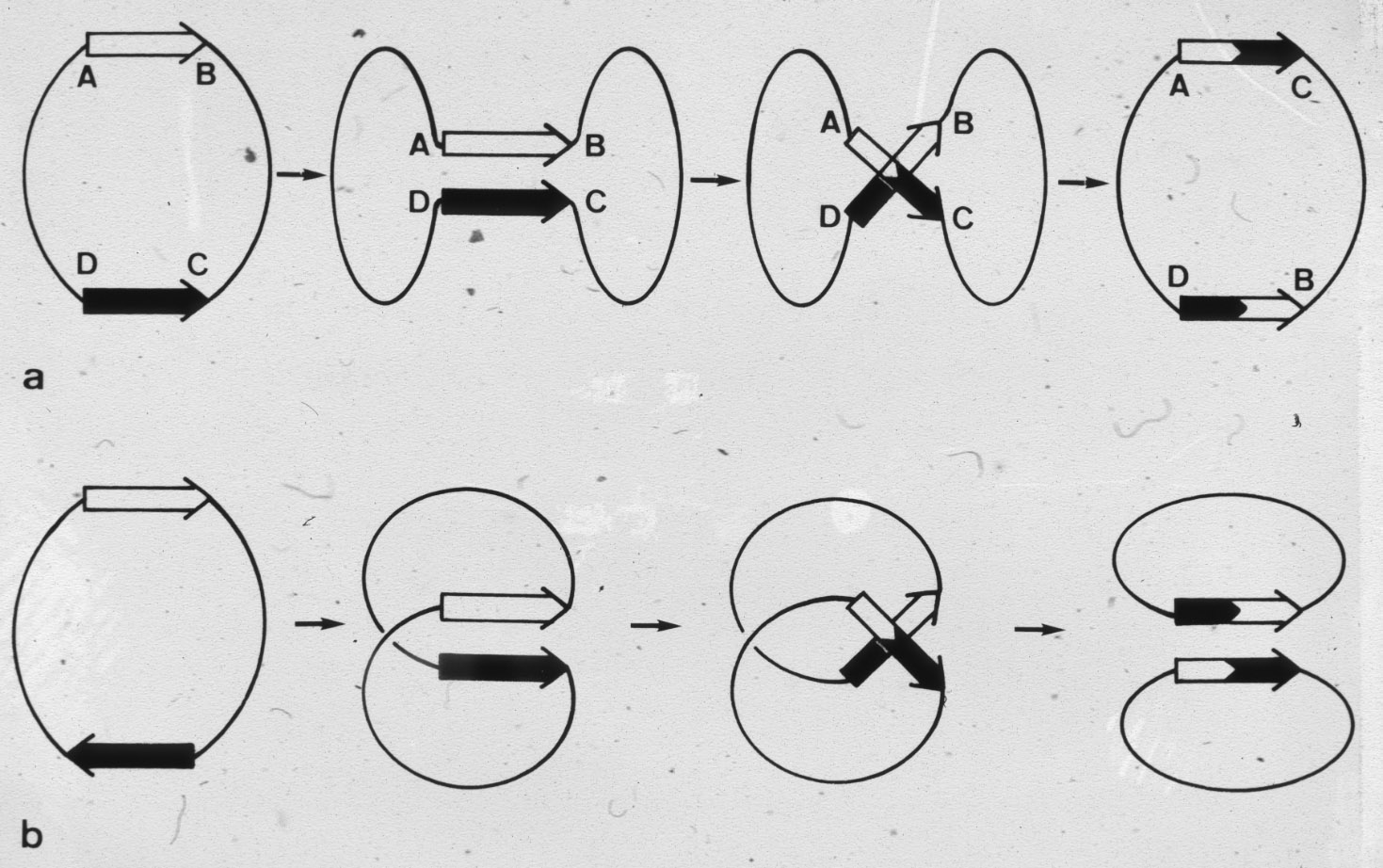 |
Figure 6: Schematic drawing of intramolecular site specific recombination reactions occurring within relaxed DNA circles, the outcome depends on the relative orientation of the sequences at which the recombination occurs. |
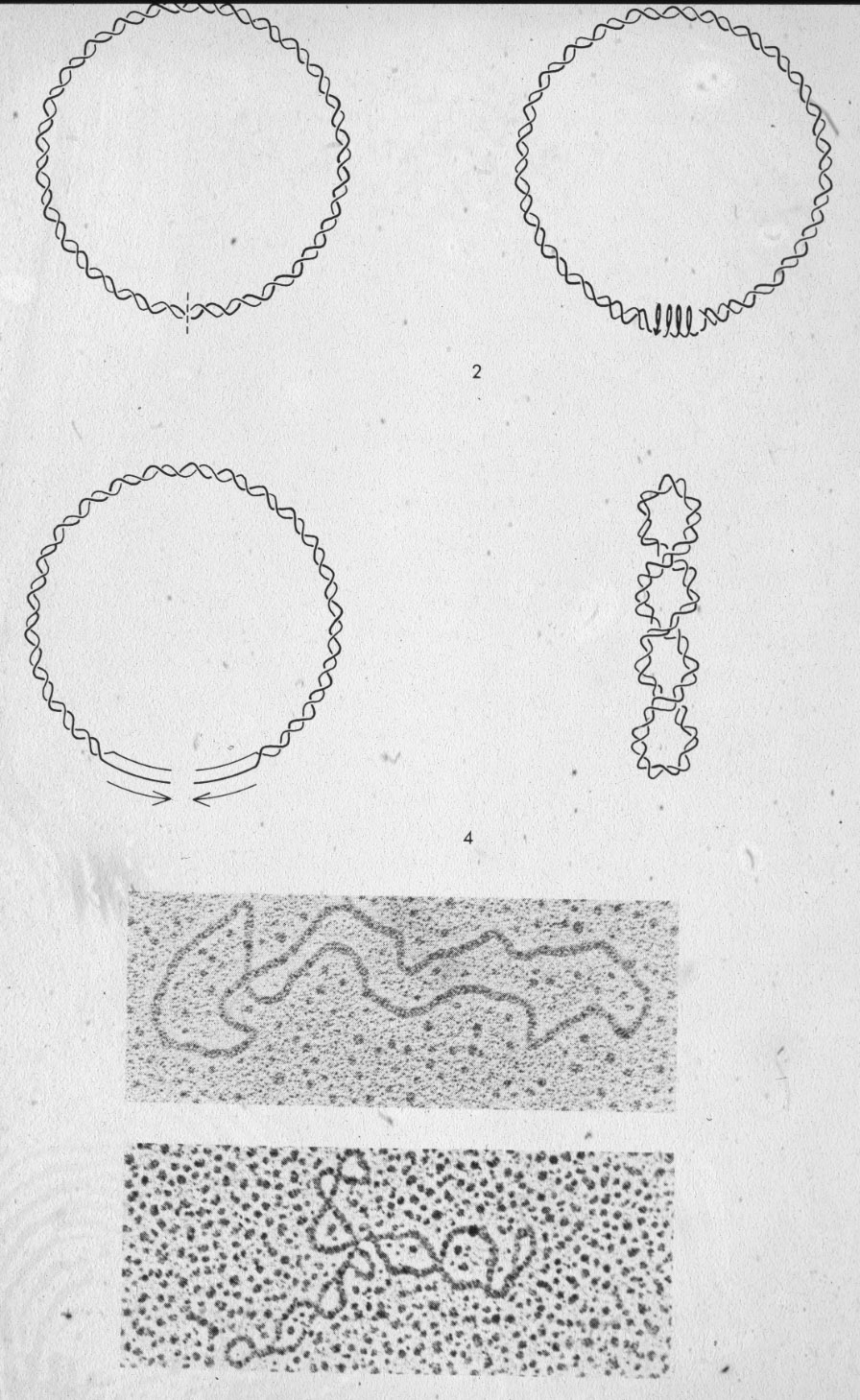 Figure 7: Naturally existing circular DNA is supercoiled.
Figure 7: Naturally existing circular DNA is supercoiled. 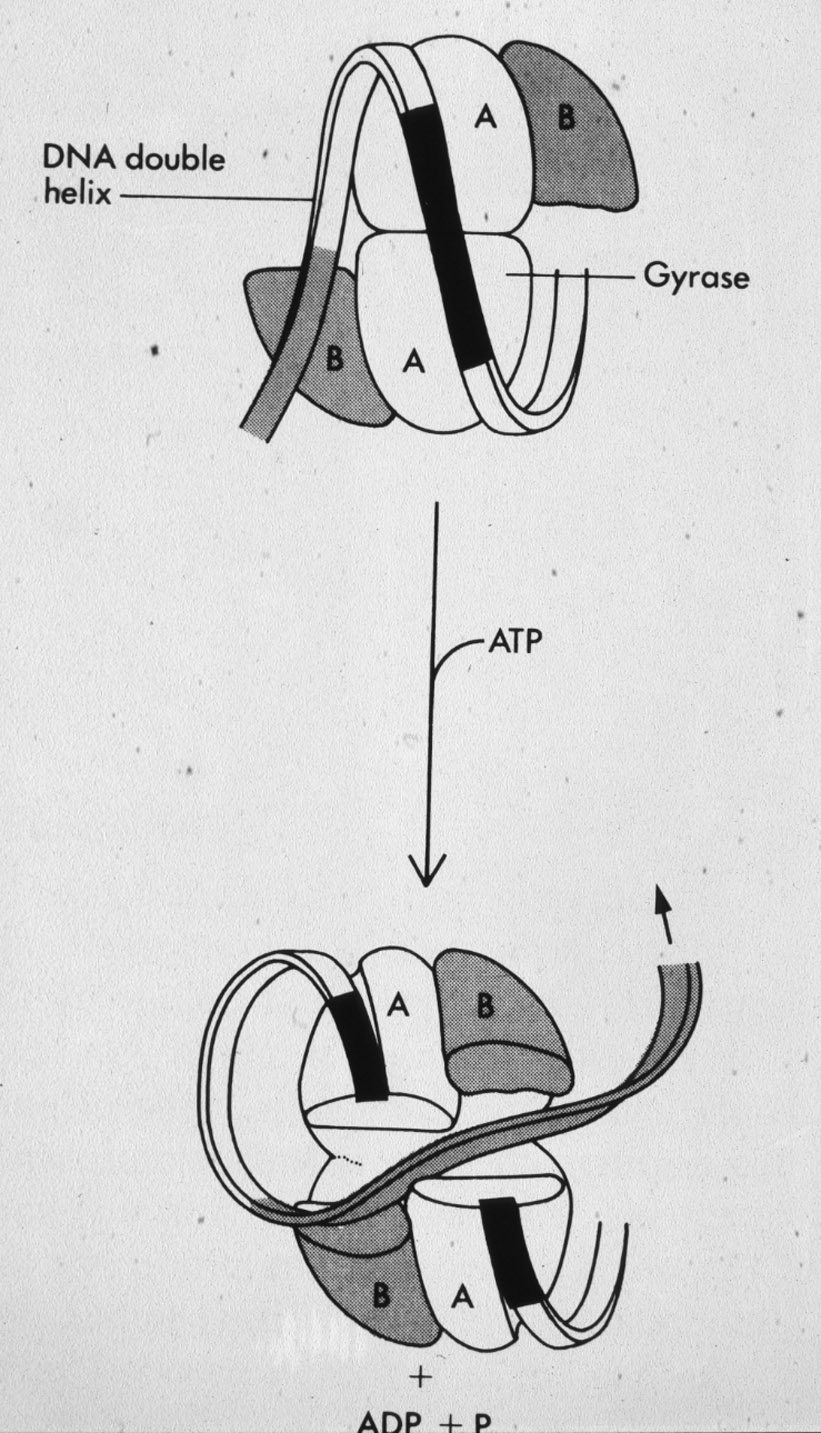 |
Figure 8: Mechanism of action of DNA gyrase that generates negative supercoiling in bacterial DNA. |
| Figure 9: Right-handed torus type of knots with various level of complexity are produced by site specific intramolecular recombination reaction mediated by phage lambda integrase (Int) when acting on recognition sequences in inverted repeat orientation. | 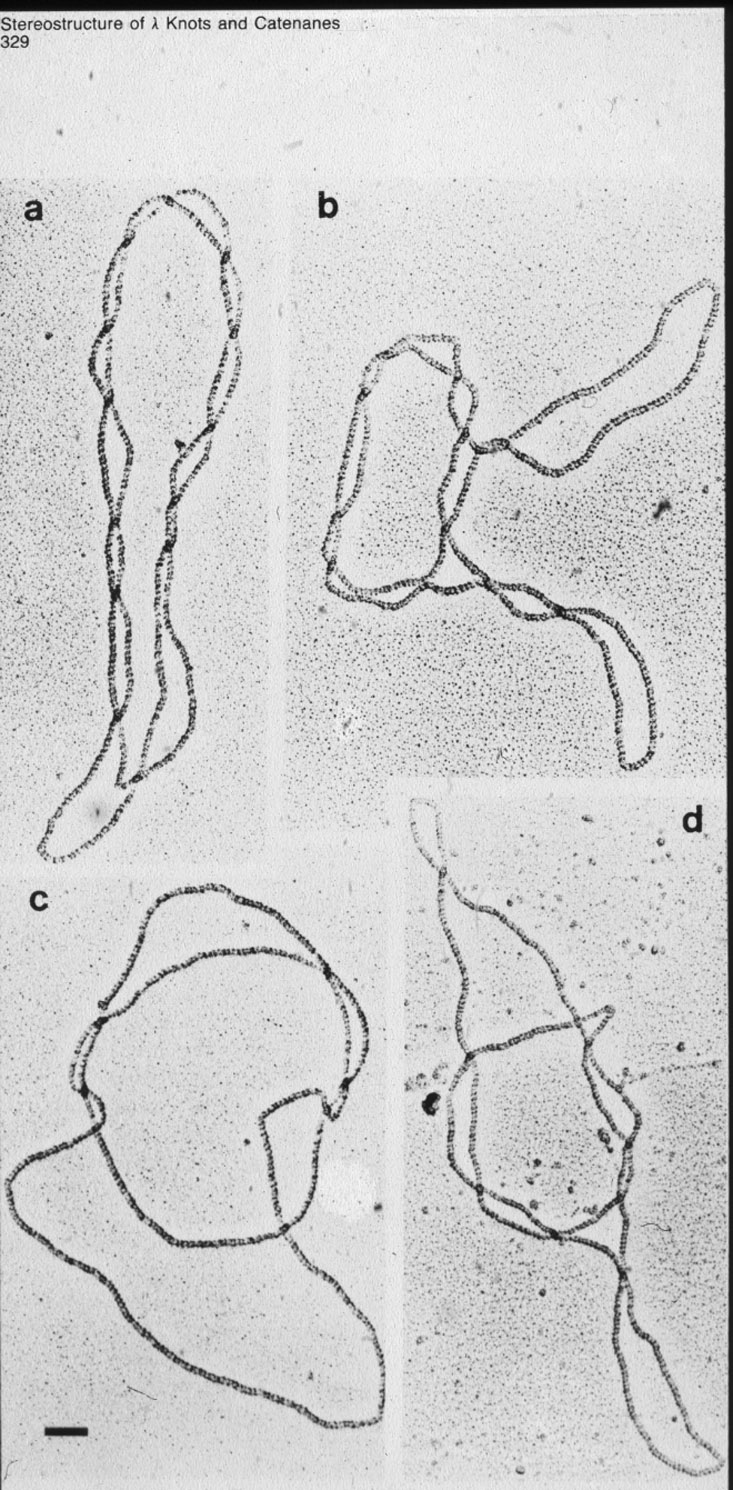 |
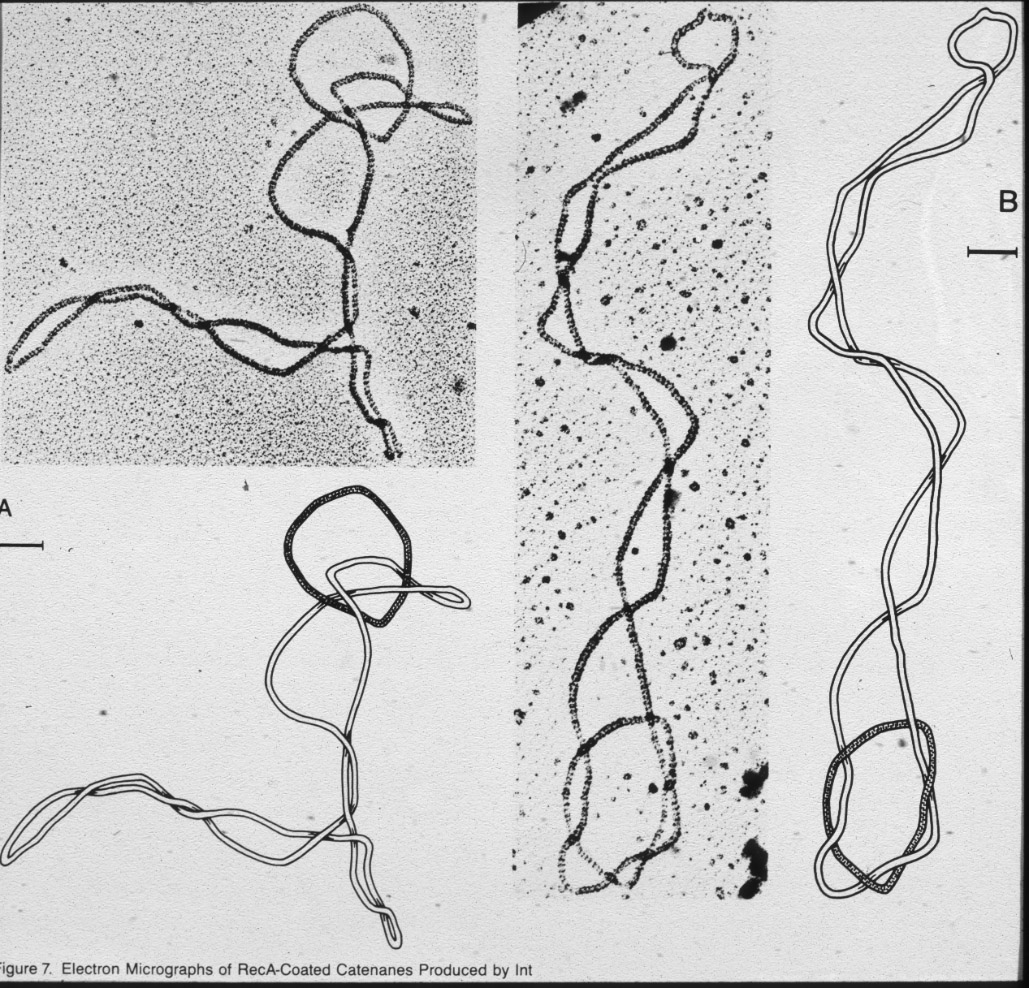 |
Figure 10: Right-handed torus type of catenanes with different level of complexity are produced by site specific intramolecular recombination reaction mediated by Int when acting on recognition sequences that are in the same orientation (see slide 6). |
| Figure 11: Right-handed torus knots and catenanes. | 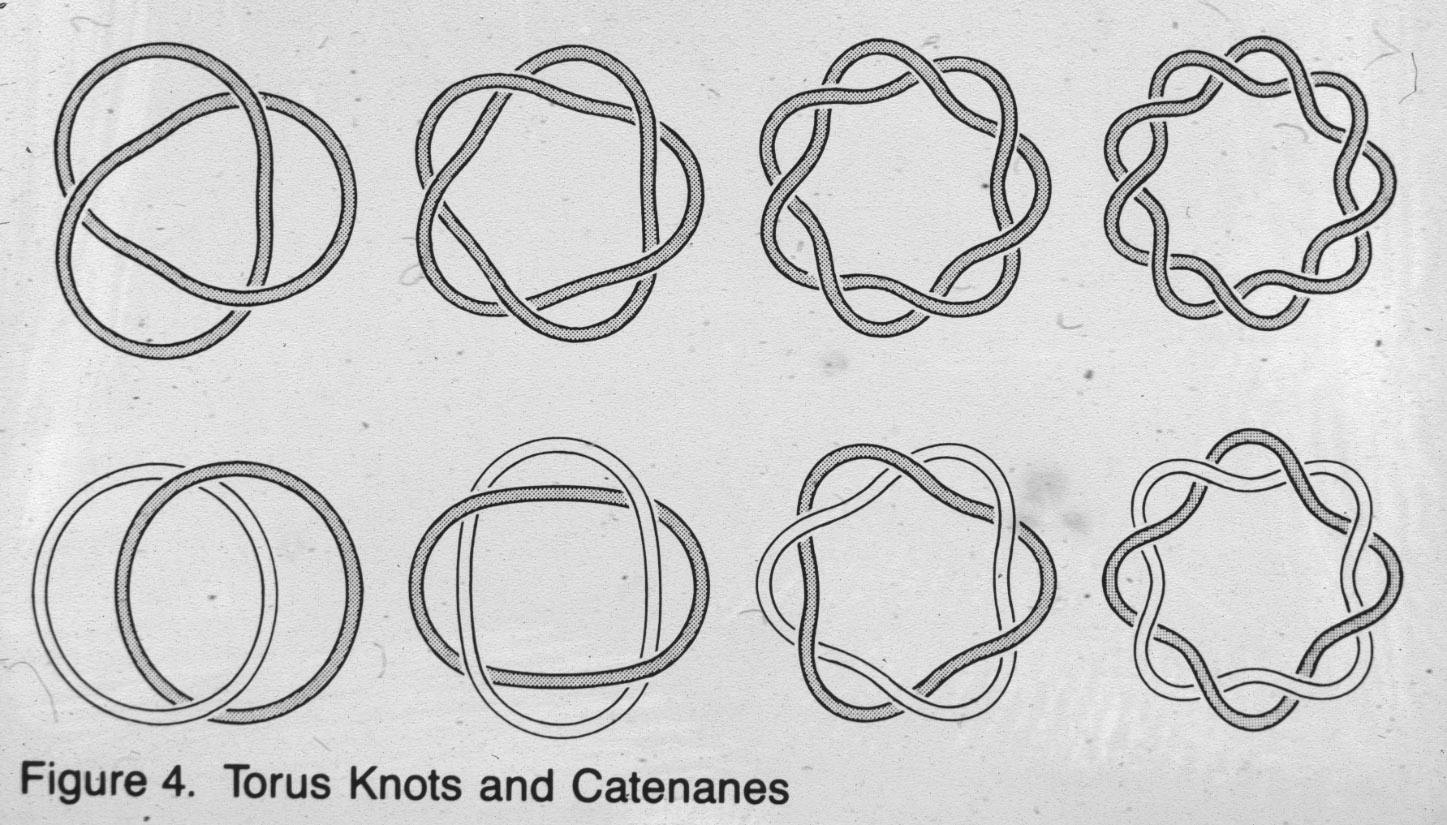 |
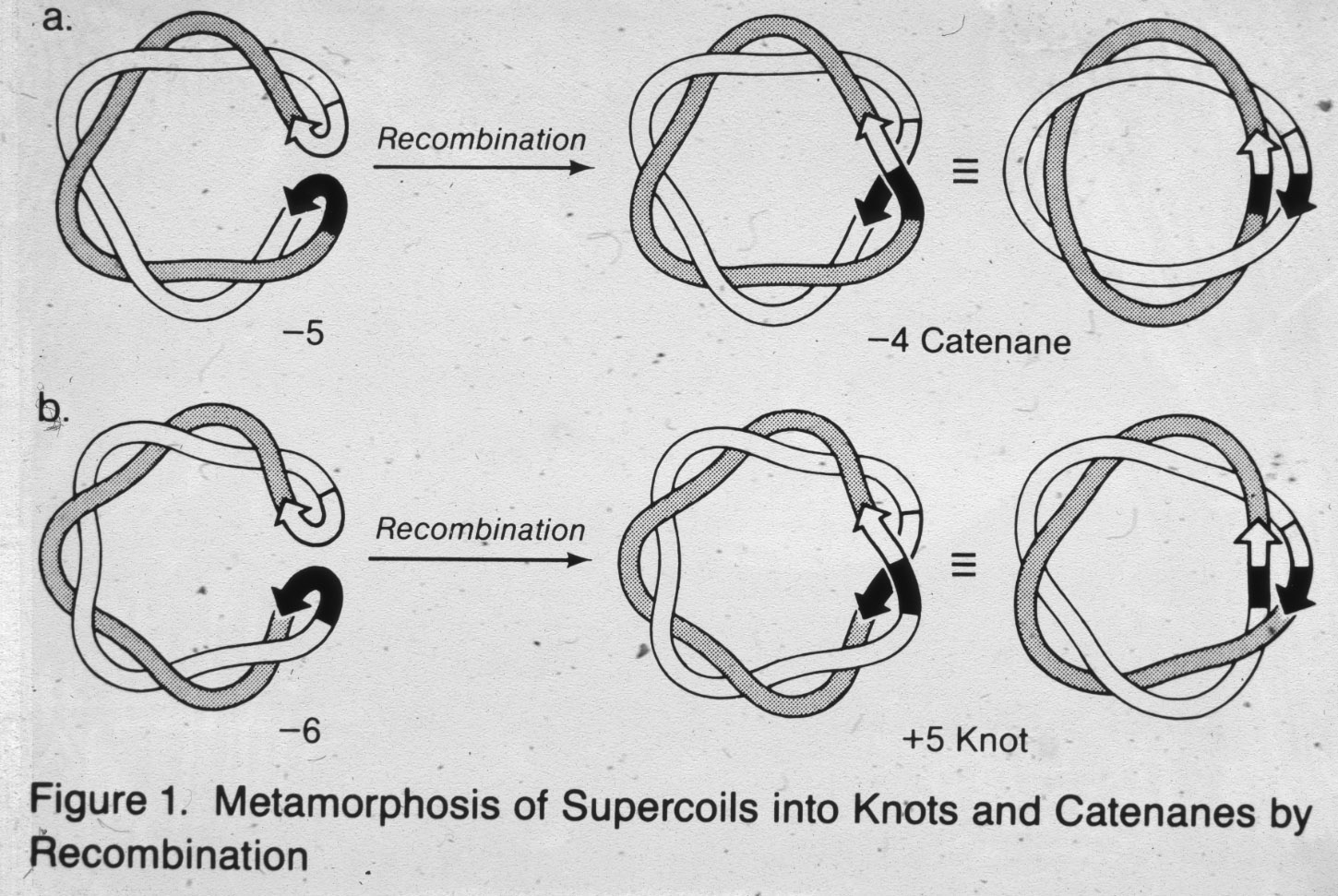 |
Figure 12: Conversion of supercoiled DNA molecules into torus knots and catenanes by action of Int protein. |
| Figure 13: Various levels of complexity of produced knots and catenanes results from thermally driven slithering type of motion within supercoiled DNA molecules. | 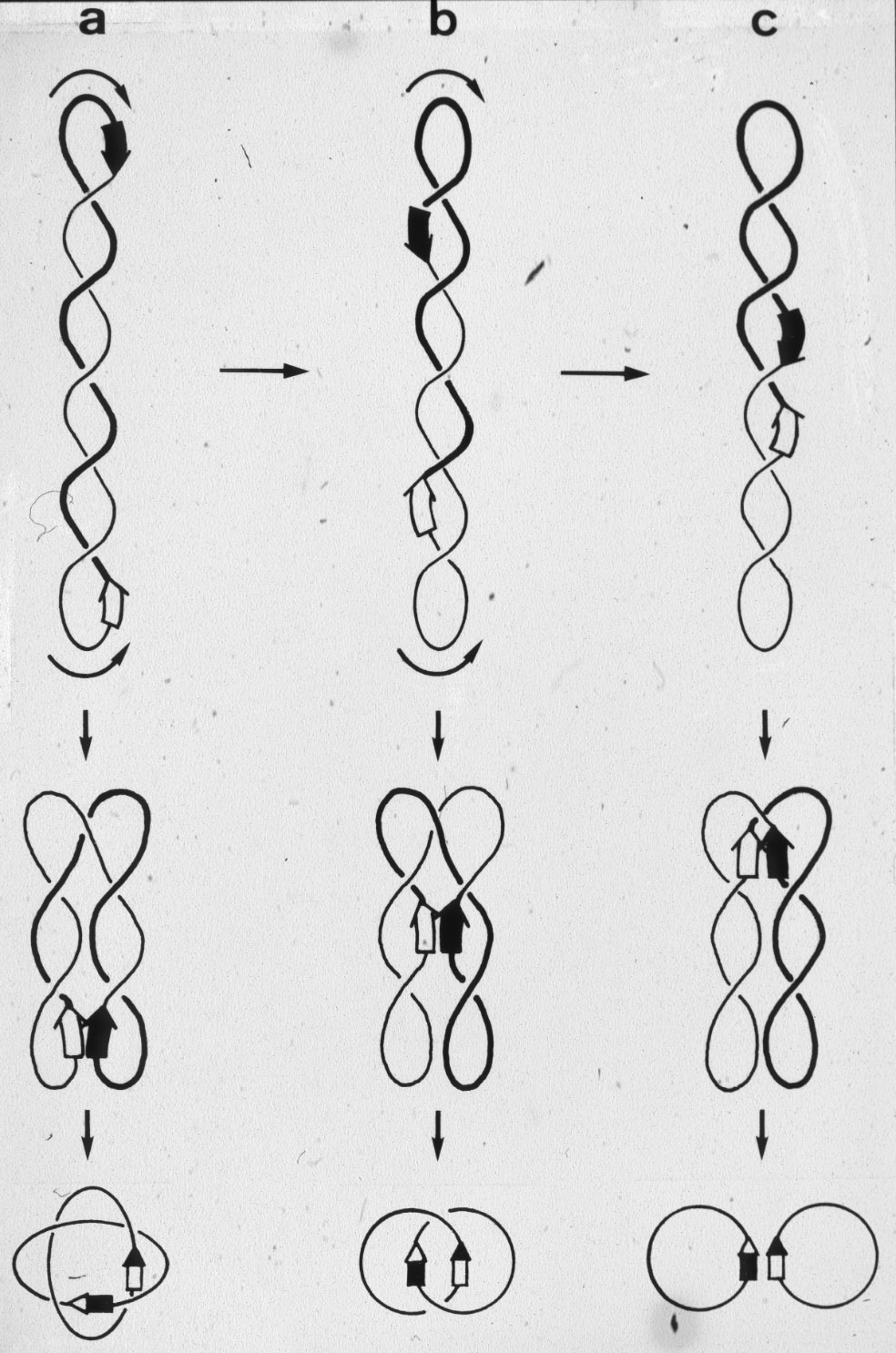 |
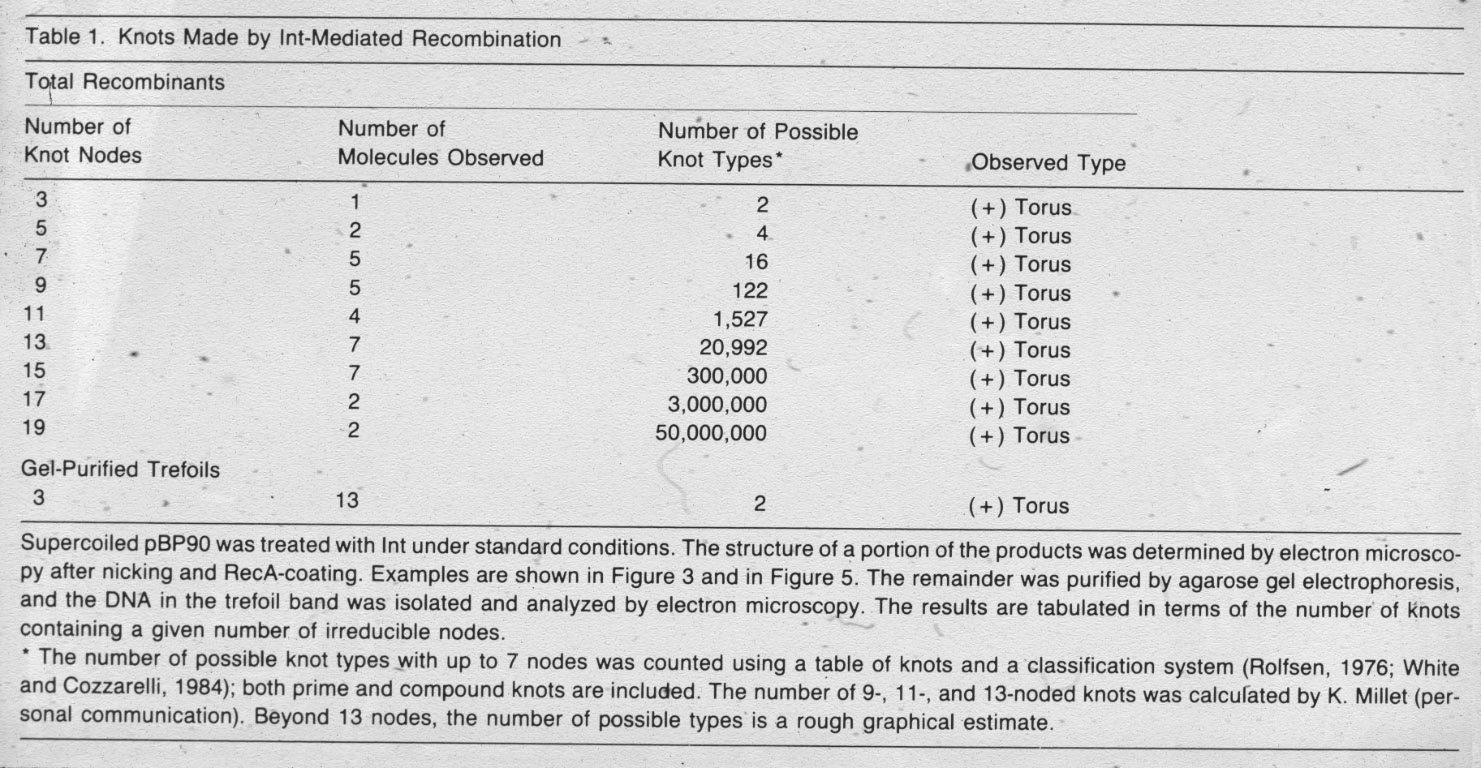 |
Figure 14: Phage lambda integrase (Int) acting on supercoiled DNA produces knots or links of only one family (torus) but with different level of complexity. |
| Figure 15: Another site specific recombination enzyme called resolvase when acting on supercoiled DNA molecules produces mainly singly linked catenanes (here analysed on a gel after relaxation of supercoils in DNA). | 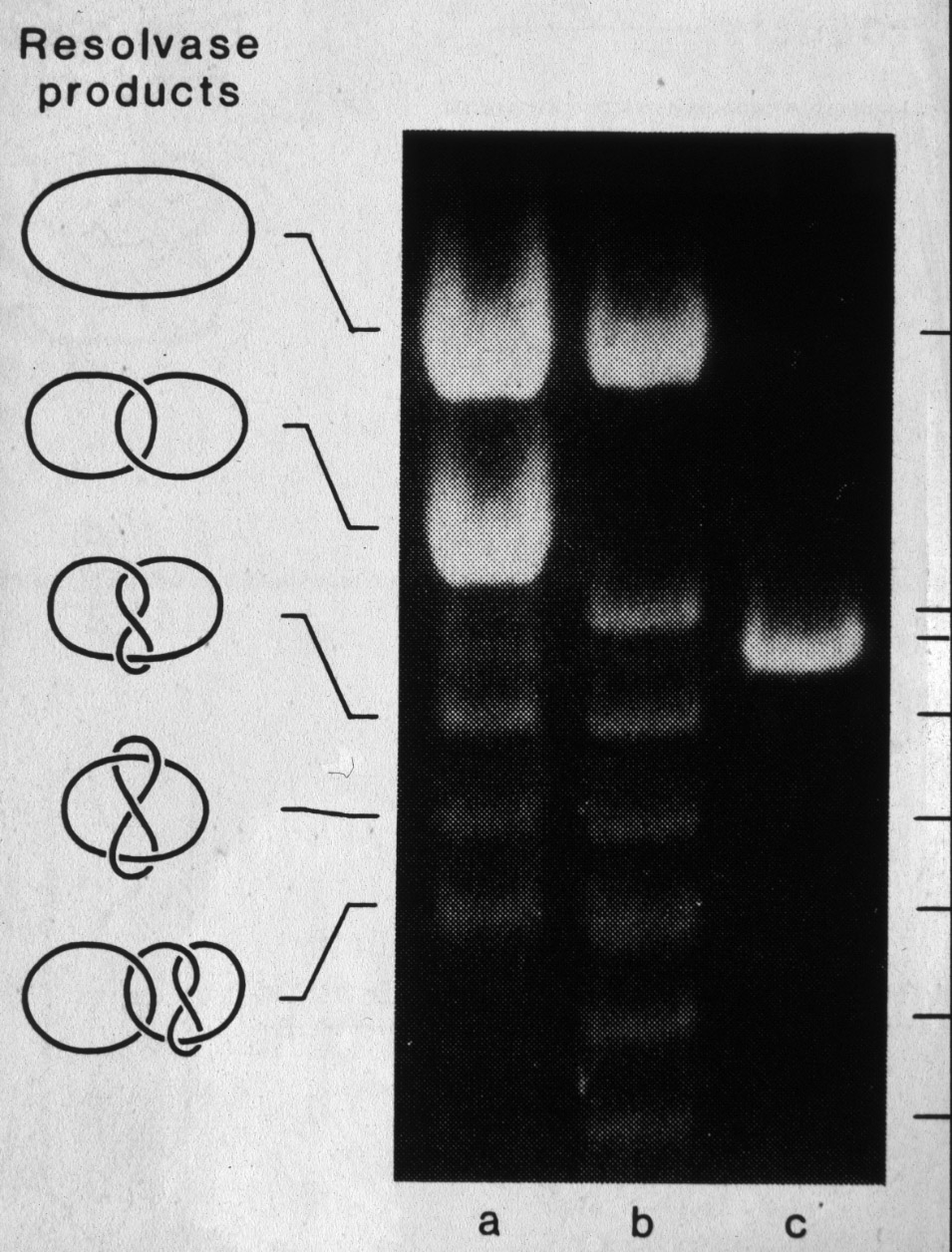 |
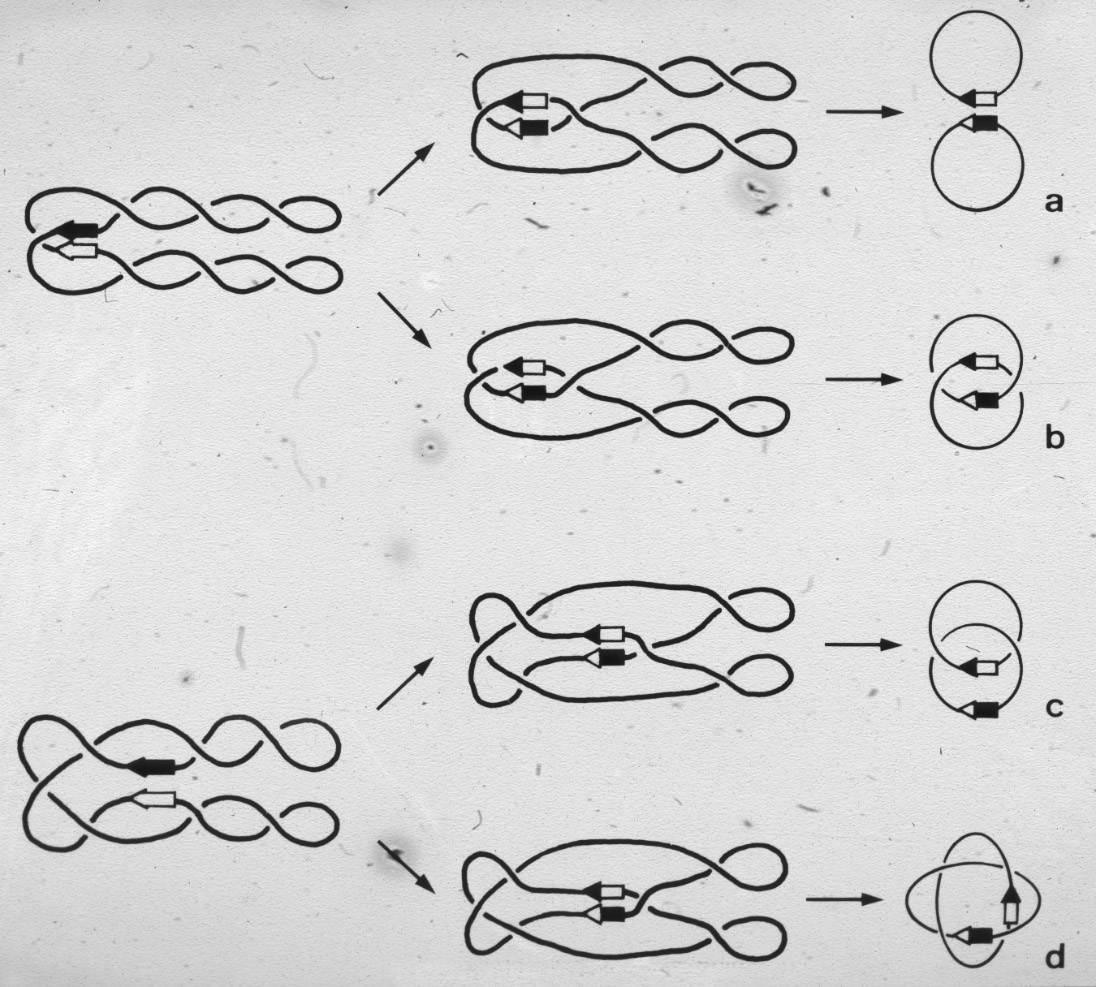 |
Figure 16: Two different possible mechanisms that could allow the resolvase to produce singly linked catenanes. |
| Figure 17: Characterisation of the major (a) and minor products of resolvase action (b,c). | 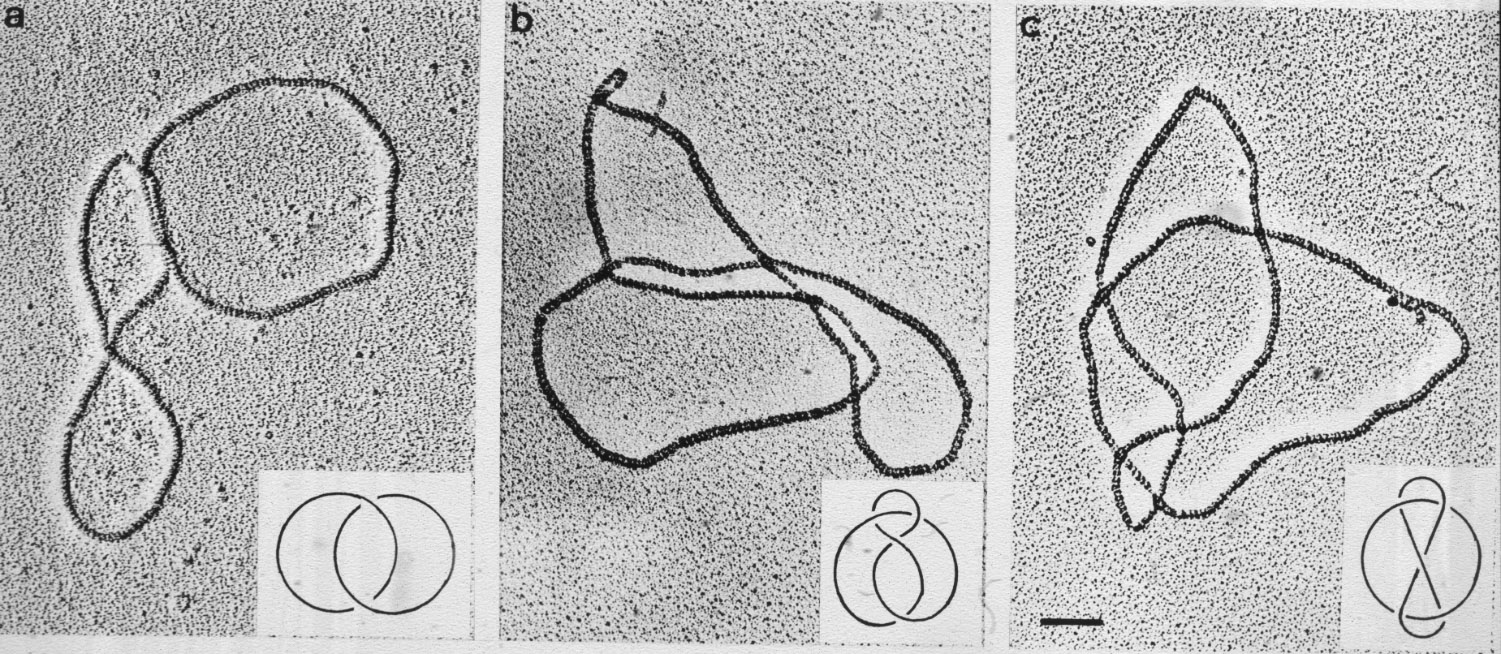 |
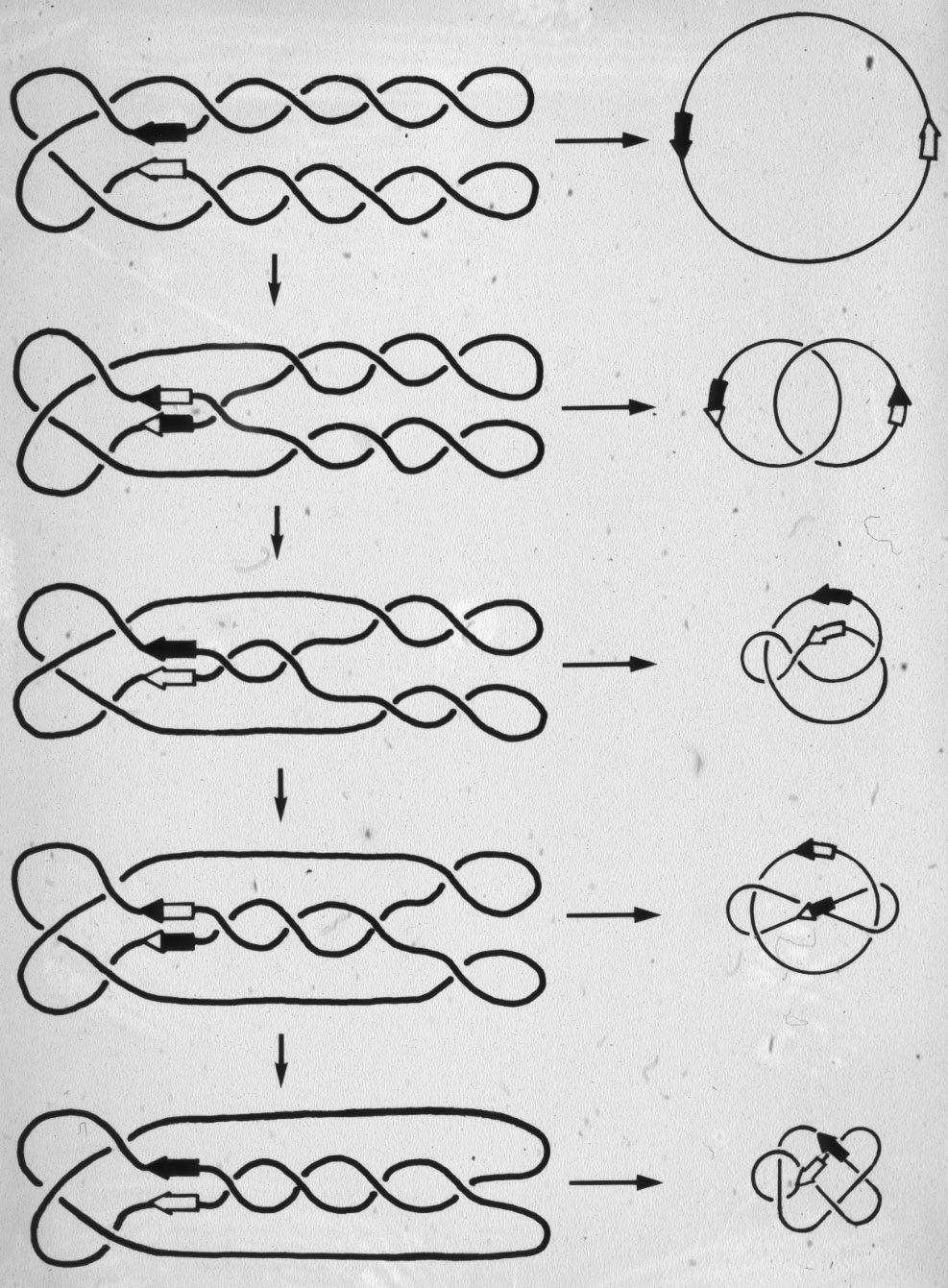 |
Figure 18: Only one mechanism of resolvase action can explain the generation of the observed minor products of the reaction. |
| Figure 19: Replication of circular DNA, topoisomerases acting on unreplicated portion of DNA avoid accumulation of interlinking in newly replicated portions. | 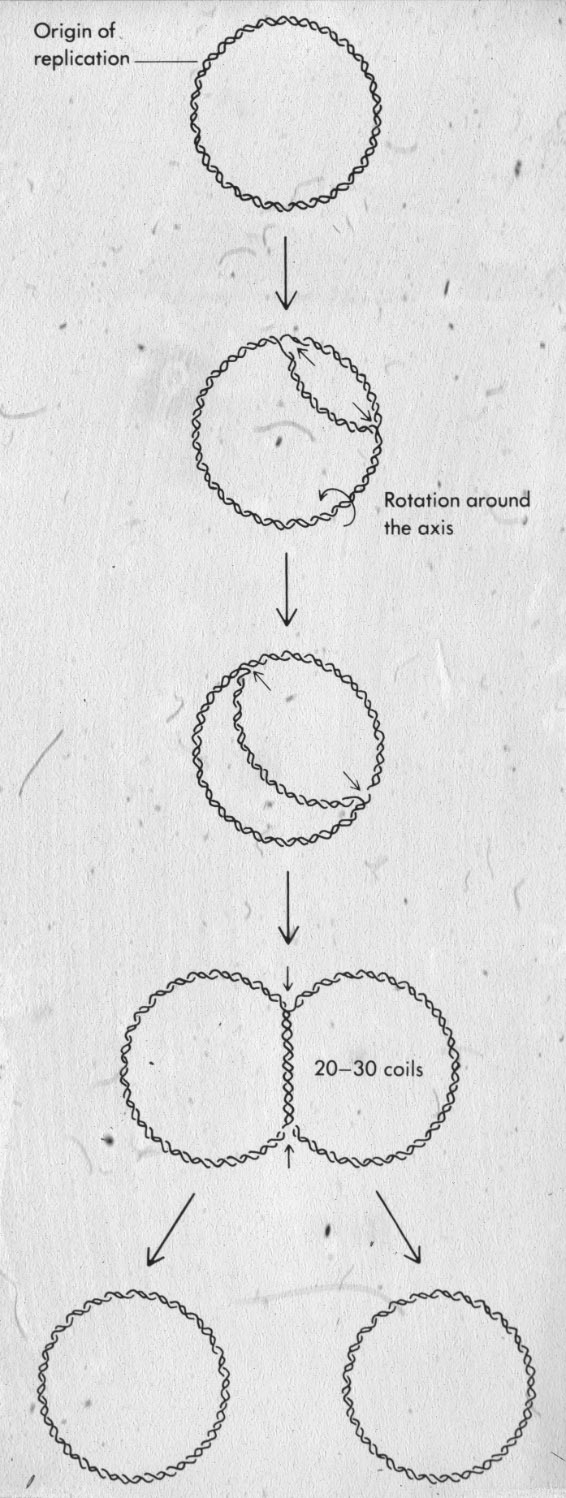 |
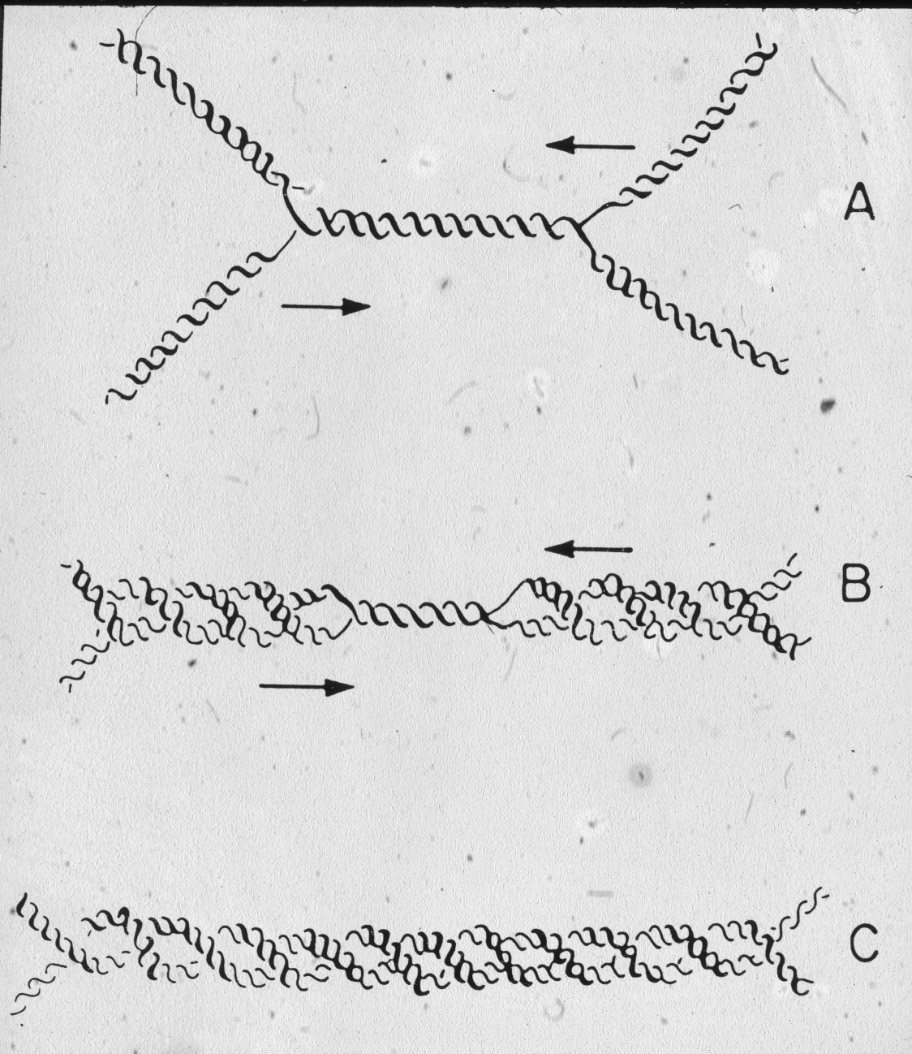 |
Figure 20: Topoisomerases require ca 200 bp for its binding, therefore when the terminal portion of ca 200 bp is replicated there is no place for the topoisomerase to act as a results right-handed interlinking of freshly replicated DNA molecules should be observed. |
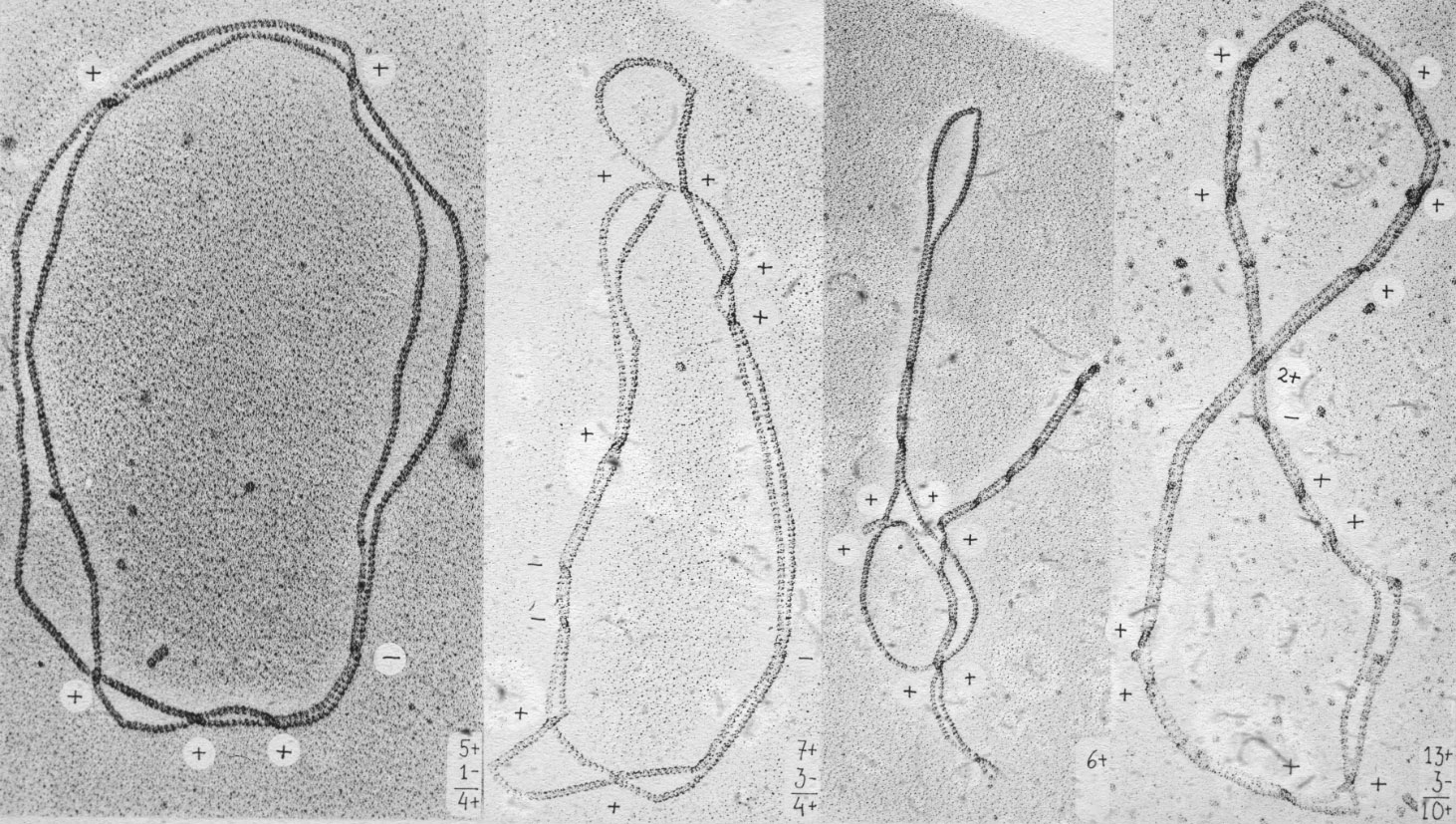 |
Figure 21: In fact such an interlinking is observed in freshly replicated circular DNA molecules. |