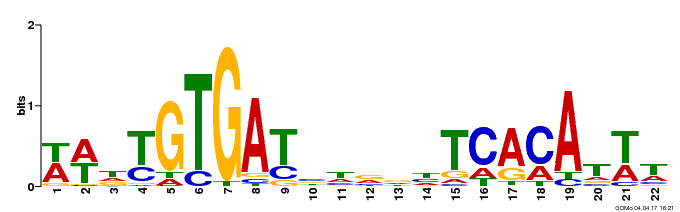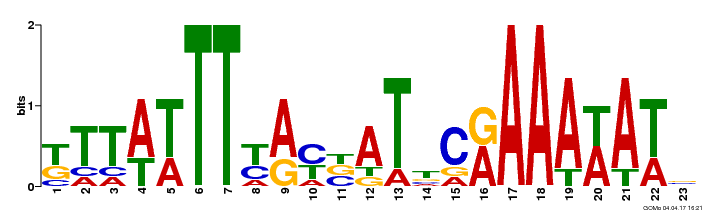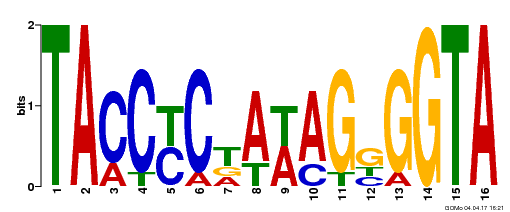| GO:0009056 |
8.869e-07 |
5.615e-07 |
4.559e-04 |
~0%
|
BP |
catabolic process |
deoC (24), iraP (31), fucA (51), fucP (52), exuT (64), uxaC (65), tdcA (71), rhaB (80), araB (143), fruB (147), ...49 more... |
| GO:0044248 |
1.341e-06 |
5.615e-07 |
4.559e-04 |
~1%
|
BP |
cellular catabolic process |
deoC (24), fucA (51), fucP (52), exuT (64), uxaC (65), tdcA (71), rhaB (80), araB (143), fruB (147), fruB (148), ...42 more... |
| GO:0008643 |
1.075e-04 |
8.422e-06 |
4.559e-03 |
~4%
|
BP |
carbohydrate transport |
fucP (52), malE (135), malK (136), ulaA (172), xylF (177), idnD (196), yjjL (840)
|
| GO:0051119 |
2.285e-04 |
2.190e-05 |
6.991e-03 |
~5%
|
MF |
sugar transmembrane transporter activity |
fucP (52), glpT (99), malE (135), malK (136), xylF (177)
|
| GO:0044275 |
2.652e-04 |
2.583e-05 |
6.991e-03 |
~7%
|
BP |
cellular carbohydrate catabolic process |
fucA (51), fucP (52), rhaB (80), araB (143), fruB (147), fruB (148), nagB (153), xylA (176), idnD (196), nanA (204), ...10 more... |
| GO:0015144 |
2.665e-04 |
2.583e-05 |
6.991e-03 |
~3%
|
MF |
carbohydrate transmembrane transporter activity |
fucP (52), glpT (99), malE (135), malK (136), ulaA (172), xylF (177), ycaI (1391)
|
| GO:0016861 |
3.954e-04 |
4.829e-05 |
1.120e-02 |
~14%
|
MF |
intramolecular oxidoreductase activity, interconverting aldoses and ketoses |
fucP (52), uxaC (65), rhaB (80), srlA (111), araB (143), nagB (153), xylA (176), hisL (637), yihU (858), rpiB (895), ...3 more... |
| GO:0016860 |
4.737e-04 |
6.569e-05 |
1.256e-02 |
~4%
|
MF |
intramolecular oxidoreductase activity |
fucP (52), uxaC (65), rhaB (80), srlA (111), araB (143), nagB (153), xylA (176), hisL (637), yihU (858), rpiB (895), ...4 more... |
| GO:0016052 |
5.421e-04 |
6.962e-05 |
1.256e-02 |
~3%
|
BP |
carbohydrate catabolic process |
fucA (51), fucP (52), rhaB (80), araB (143), fruB (147), fruB (148), nagB (153), ybjQ (159), xylA (176), idnD (196), ...14 more... |
| GO:0009057 |
5.850e-04 |
7.805e-05 |
1.268e-02 |
~2%
|
BP |
macromolecule catabolic process |
iraP (31), fucA (51), fucP (52), rhaB (80), araB (143), fruB (147), fruB (148), nagB (153), ybjQ (159), xylA (176), ...20 more... |
| GO:0016853 |
7.322e-04 |
1.067e-04 |
1.575e-02 |
~1%
|
MF |
isomerase activity |
fucP (52), uxaC (65), yqcC (78), rhaB (80), srlA (111), araB (143), nagB (153), xylA (176), nanA (204), nanC (238), ...27 more... |
| GO:0044265 |
8.893e-04 |
1.364e-04 |
1.847e-02 |
~6%
|
BP |
cellular macromolecule catabolic process |
fucA (51), fucP (52), rhaB (80), araB (143), fruB (147), fruB (148), nagB (153), xylA (176), idnD (196), nanA (204), ...13 more... |
| GO:0019321 |
1.272e-03 |
2.049e-04 |
2.560e-02 |
~16%
|
BP |
pentose metabolic process |
fucA (51), fucP (52), rhaB (80), araB (143), xylA (176), rbsD (365), aldA (1059), yiaK (1950)
|