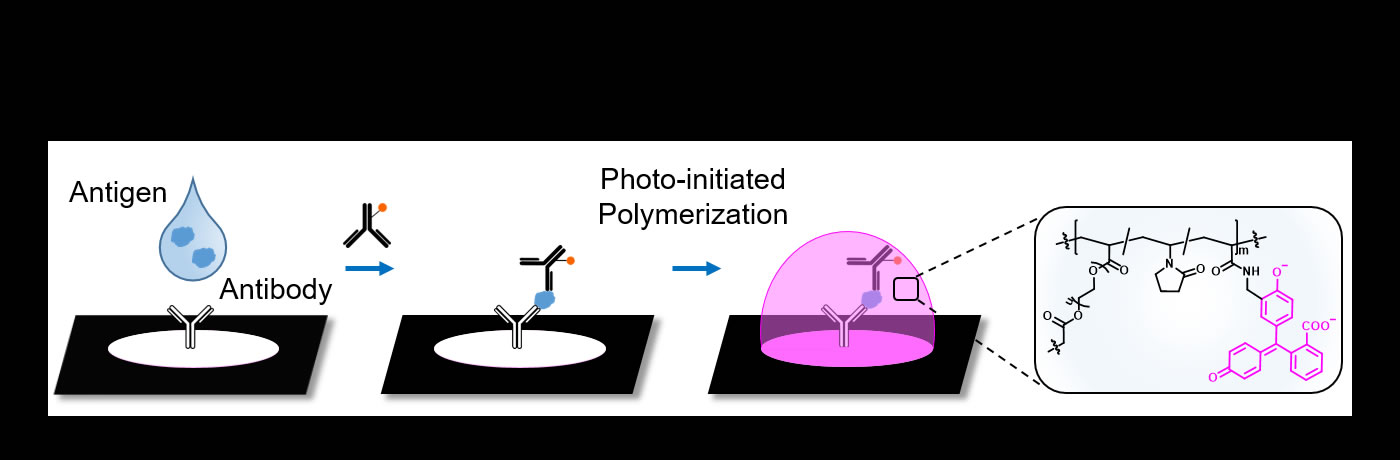
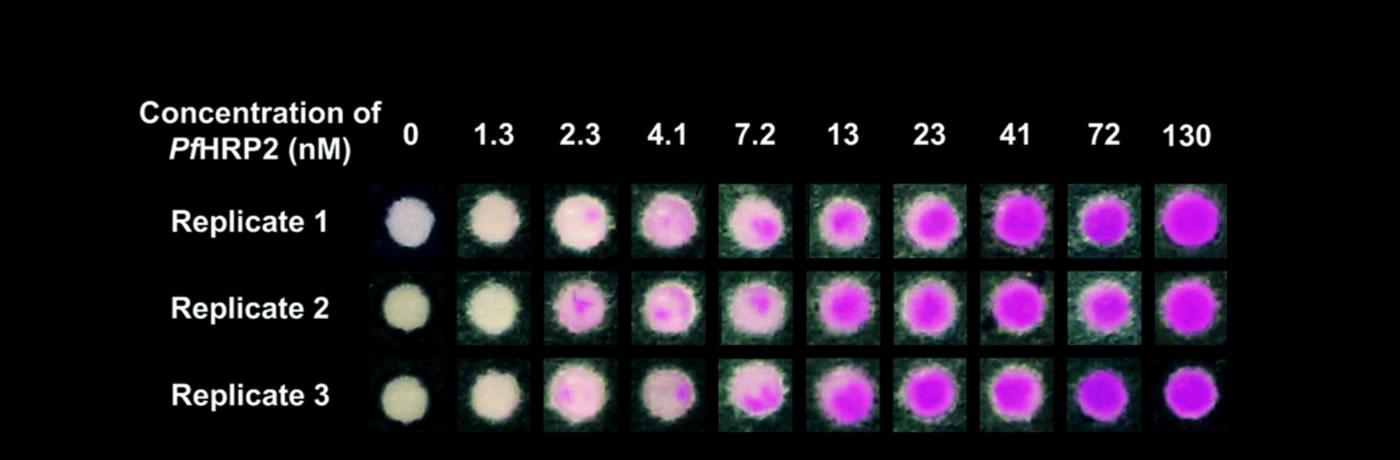
Research Overview
Our efforts focus on engineering biomolecular systems to detect and treat disease in new ways. We use the principles of engineering design to support and extend the practice of evidence-based diagnosis and selection of therapy.
Engineering design starts with interviewing intended users to formulate a quantitative problem statement and to understand the context and constraints for a new medical test. In the area of infectious disease, proteins in bodily fluids can indicate malaria or tuberculosis. The protein identity, quantity, and bodily fluid varies with the disease. In cancer, particular epigenetic and post-translational protein modifications can predict which therapies are likely to be effective against an individual tumor. We use an understanding of thermodynamics, kinetics, and transport phenomena to design medical tests that simultaneously meet design criteria for analytical performance, assay time, cost, robustness, and infrastructural requirements. We iteratively test prototypes with clinical collaborators to assess and improve real-world utility.