
Although microfluidic chips have demonstrated basic functionality for single applications, performing varied and complex experiments on a single device is still technically challenging. While many groups have implemented control software to drive the pumps, valves, and electrodes used to manipulate fluids in microfluidic devices, a new level of programmability is needed for end users to orchestrate their own unique experiments on a given device. With this goal in mind, we have been collaborating with Prof. Saman Amarasinghe's group at MIT on the development of high-level software libraries to enable large, complex biology experiments to be easily executed on microfluidic chips designed by the end-user. The source code, named BioStream, is freely available. E-mail Bill Thies if interested (thies@mit.edu).
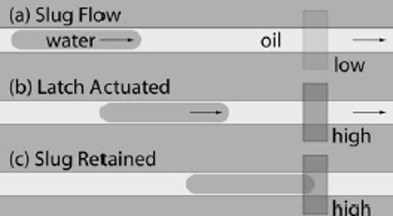
As a proof-of-concept platform for the software, we developed a two-phase microfluidic chip (water-in-oil), where the water droplets are of a unit length defined by latched chambers. a novel ‘‘microfluidic latch’’ is used to precisely align a sample after it has been transported a long distance through the flow channels. To demonstrate the scalability of the approach, this paper introduces a ‘‘generalpurpose’’ microfluidic chip containing a rotary mixer and addressable storage cells. The system is general purpose in that all operations on the chip operate in terms of unit-sized aqueous samples; using the underlying mechanisms for sample transport and storage, additional sensors and
actuators can be integrated in a scalable manner. A novel high-level software library allows users to specify experiments in terms of variables (i.e., fluids) and operations (i.e., mixes) without the need for detailed knowledge about the underlying device architecture. This research represents a first step to provide a programmable interface to the microfluidic realm, with the aim of enabling a new level of scalability and flexibility for lab-on-a-chip experiments (Ref 3).
  |
We are currently applying the sample automation technology to a number of projects in the laboratory, including a platform to study cell signalling and biochemical reaction pathways with Prof. Jeremy Guanwardena at Harvard Medical School and a system for monitoring in vitro fertilization.
Present Post-Doctoral Fellows
Present Graduate Students
 |
David Craig (Masters Student) |
 |
J.P. Urbanski (Doctoral Student) |
 |
Bill Thies (Doctoral Student - Amarasinghe Group) |
Present Undergraduate Students
Visitors
 |
Natalie Andrew (Post-doctoral fellow - Gunawardena Group) |
Alumni
Collaborators
Professor Saman Amarasinghe, Dept. of Electrical Engineering and Computer Science, M.I.T.
Professor Jeremy Gunawardena, Dept. of Systems Biology, Harvard University
Sponsors
National Science Foundation (CCF-0541319)
Links
Programmable Microfluidics Bill Thies website
Refereed Publications and Conference Proceedings
1. Todd Thorsen. Microfluidic Logic - Addressing Complexity at the Microscale. ASME IMECE (2003)
2. W. Thies, J.P. Urbanksi, M. Cooper, D. Wentzlaff, T. Thorsen, and S. Amarasinghe. Programmable Microfluidics. Eleventh International Conference on Architectural Support for Programming Languages and Operating Systems - Wild and Crazy Ideas Session (2004)
3. J.P. Urbanski, W. Thies, C. Rhodes, S. Amarasinghe, and T. Thorsen. Digital microfluidics using soft lithography. Lab Chip 6: 96-104 (2006) (PDF)
4. W. Thies, J.P. Urbanski, T. Thorsen, and S. Amarasinghe, Abstraction Layers for Scalable Microfluidic Biocomputers. 12th International Meeting on DNA Computing Seoul, Korea (2006). Published in Lec. Notes Comput. Science , 4287: 308-323 (2006) (PDF)
5. W. Thies, J.P. Urbanski, T. Thorsen, and S. Amarasinghe. Abstraction Layers for Scalable Microfluidic Biocomputing. Natural Computing , DOI 10.1007/s11047-006-9032-6 (2007) (PDF)