
|

|
| |
||||||||||||||||||||
| Main | Publications | Journals | Members | Databases | ||||||||||||||||
Laboratory of Michael B. Yaffe, M.D., Ph.D.
at the MIT Center for Cancer Research

|
Research Overview
Regulation of protein-protein interactions and signal transduction pathways by protein and lipid phosphorylation. Structure and function of modular signaling domains. Design of bioinformatics tools for proteomic analysis.
The goal of our research is to understand how protein phosphorylation controls progression through the cell cycle at the molecular level and how defects in phosphorylation bypass normal cell cycle checkpoints and lead to human cancer. We also study how protein and lipid phosphorylation controls the inflammatory response in phagocytic cells.
|
Research Summary
Cells activate complex signaling pathways in response to stresses and injuries such as DNA damage, hypoxia, and bacterial or viral infection. These pathways control cell cycle progression, coordinately regulate patterns of gene expression and/or initiate programmed cell death by activating protein serine/threonine kinases that phosphorylate critical downstream targets. Mutations in signaling pathways that normally respond to either DNA damage or to disruption of the mitotic spindle, for example, play critical roles in the genesis of most human cancers. Similarly, hypoxia and infection cause dysregulation of cell signaling pathways in phagocytic cells, causing tissue damage in auto-inflammatory diseases and multiple organ failure in sepsis.
How are these signaling pathways assembled? What are the key molecules involved? How does their phosphorylation on serine and threonine residues regulate protein-protein interactions to control the cell cycle in epithelial cells or regulate the production of inflammatory mediators by phagocytic cells? Our lab uses a broad proteomics approach to decode how these cell signaling pathways are "wired" using bioinformatics, combinatorial chemistry, cell biology, physical biochemistry, structural biology and molecular genetics.
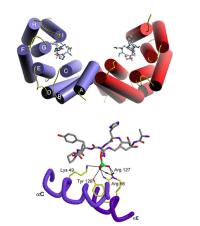
Modular domains that mediate the phosphoserine/threonine-dependent assembly of signaling complexes. A major clue to understanding cell signaling was the discovery that many molecules contain discrete modular signaling domains such as SH2, SH3 and PDZ domains, arranged in a combinatorial fashion. SH2 and PTB domains bind to phosphotyrosine- containing sequence motifs, but the fundamental mechanisms underlying phosphoserine/phosphothreonine signaling have remained poorly understood. Very recently, a subset of new modules, including 14-3-3 proteins, FHA domains and WW domains, were found to bind specifically to short phosphoserine or phosphothreonine-containing amino acid sequences within proteins. These pSer/pThr-binding modules integrate signals from upstream protein Ser/Thr kinases to control the actions of downstream effector molecules, controlling cell cycle checkpoints and activating patterns of gene transcription. We are using phosphoserine- and phosphothreonine-oriented peptide libraries to elucidate the specific sequence motifs recognized by each of these modules and have developed bioinformatics algorithms that use this information to identify likely interacting proteins within the mouse, human and yeast proteomes. In parallel with this combinatorial chemistry/computational approach, we are using high-density protein arrays and expression libraries to identify specific interacting proteins that play critical roles in establishing cell cycle checkpoints that respond to DNA damage or disruption of the mitotic spindle. In addition, we are developing a novel library-against-library screening approach which should reveal additional phosphoserine/threonine-binding modules that have not yet been identified. In a long-standing joint effort with the Drs. Stephen Smerdon and Steven Gamblin in the Division of Protein Structure at the National Institute of Medical Research in London we are elucidating the structural basis of pSer/pThr binding for 14-3-3 and FHA domains by X-ray crystallography. We hope to be able to use this information to design specific chemical inhibitors that can serve as research tools for cell biology and may act as lead compounds for drug design in the treatment of human cancer.
Neutrophil Signaling in Inflammation. Neutrophils are professional phagocytic cells that constitute the first line of defense against infection. In response to cytokines and bacterial products, neutrophils generate reactive oxygen species to kill pathogens. Excessive amounts of reactive oxygen products, however, damage adjacent host tissues, causing the auto-inflammation seen in diseases like rheumatoid arthritis and the lung and kidney failure observed in patients with fulminant infections, like gram negative sepsis and Legionnaire's disease. The production of reactive oxygen by the NADPH oxidase complex must therefore be tightly controlled.
The NADPH oxidase consists of at least 6 subunits - 4 cytosolic proteins p47phox, p67phox, p40phox and Rac, and 2 membrane proteins - p22phox and gp91phox. The cytosolic proteins contain SH3 domains, PX domains and TPR repeats, through which the cytosolic subunits bind to the membrane subunits in response to activation of protein Ser/Thr kinases and lipid kinases. Most of the details of how this occurs, as well as the relevant kinase signaling pathways, are still unknown. We have recently found that cytokines like TNFa can "prime" neutrophils to generate excessive amounts of reactive oxygen and cause multiple organ failure through a process that involves the p38 MAPK pathway. Using peptide and protein libraries, bioinformatics, ligand screening approaches and a novel cell based assay using human neutrophils, we are decoding the sequence motifs recognized by the modular signaling domains in p47phox, p40phox and p67phox, and the molecular targets of the relevant Ser/Thr kinases and lipid kinases involved. We have recently discovered that the PX domains of p47phox and p40phox recognize specific phosphoinositol lipids generated by the signaling enzyme PI 3-kinase, providing a new conceptual model for how the cytosolic components migrate to the membrane upon activation. Using green fluorescent protein fused to domains from the cytosolic subunits of the NADPH oxidase, we are attempting to analyze the process of oxidase assembly in real time in phagocytic cells. Together with Dr. Katrin Rittinger at the National Institute of Medical Research in London, we are pursuing the structure of the NADPH oxidase using X-ray crystallography.
| Home | Publications | Journals | Members | Databases |