Aggregation Intermediates
Margaret Speed
The formation of insoluble aggregates due to protein misfolding is
a phenomenon associated with various human diseases, inclusion body
formation, and in vitro aggregation (Marston, 1986; Mitraki
and King, 1989; DeBernardez-Clark and Georgiou, 1991; Wetzel, 1994).
In vivo, this folding problem frequently arises with
heterologous proteins overexpressed in E. coli, which form inclusion
bodies, or amorphous aggregates within the cell. The analogous
aggregation problem occurs in vitro via a similar association
mechanism (Zeittmeissl, Rudolph, and Jaenicke, 1979; Colon and Kelly,
1992; Mitraki et al., 1991). Non-native multimerization subsequently
leads irreversibly to the formation of large aggregates or inclusion
bodies. A frequently employed strategy for minimizing in vitro
aggregation is to refold protein under dilute conditions. The
drawback of this practice is that it dilutes the final product, and
the strategy does not work for oligomeric proteins that cannot
assemble efficiently under dilute conditions (Jaenicke and Rudolph,
1986; Teschke and King, 1993). Therefore, further study of the
aggregation reaction is essential in designing effective refolding
strategies.
To study the polymerization process, nondenaturing polyacrylamide
gel electrophoresis (PAGE) has been used to trap a sequential ladder
of multimeric intermediates in the in vitro aggregation of P22
tailspike polypeptide chains. The aggregation intermediates 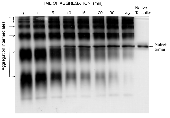 polymerized by noncovalent association of a monomeric folding
intermediate rather than aggregation of nativelike trimeric species.
These multimers were sensitive to SDS and protease unlike the
thermostable native trimer. Using a modified nondenaturing Western
blot procedure, aggregation intermediates were determined to have
nonnative epitopes in common with productive folding intermediates.
The aggregate epitopes were found on folding and aggregation
intermediates but not on denatured or native tailspike protein. The
nonnative epitope on the folding and aggregation intermediates was
located on the partially folded N-terminus, and this structural
information has implications on the aggregation mechanism.
polymerized by noncovalent association of a monomeric folding
intermediate rather than aggregation of nativelike trimeric species.
These multimers were sensitive to SDS and protease unlike the
thermostable native trimer. Using a modified nondenaturing Western
blot procedure, aggregation intermediates were determined to have
nonnative epitopes in common with productive folding intermediates.
The aggregate epitopes were found on folding and aggregation
intermediates but not on denatured or native tailspike protein. The
nonnative epitope on the folding and aggregation intermediates was
located on the partially folded N-terminus, and this structural
information has implications on the aggregation mechanism.
Although the understanding of the mechanism of protein folding and
aggregation is somewhat limited, the ability to influence the kinetic
competition between productive folding and aggregation is a
requirement for improving the recovery of active protein expressed in
a heterologous host cell (Mitraki & King, 1989). In certain
polymerization processes, such as crystallization and amyloid fibril
formation, the initial nucleation complex forms slowly, but rapid
polymerization can occur once the aggregate reaches a certain
critical size. Biological systems that display nucleation-growth
kinetics include the polymerization of sickle cell hemoglobin
(Ferrone et al., 1980), amyloid fibril formation (Jarrett &
Lansbury, 1992), actin polymerization (Tobacman and Korn, 1983),
bacterial flagella assembly (Asakura et al., 1964), and collagen
refolding (Harrington & Karr, 1970). Many biological self
assembly occur via a sequential polymerization mechanism, in which
monomeric subunits add sequentially to the growing fibril.
Two-dimensional PAGE data show that aggregation occurs by multimeric
polymerization, in which two multimers of any size can associate to
form a larger aggregate. This mechanism allows the generation of
large aggregates even after the pool of monomers has been depleted.
The aggregation intermediates are not in rapid equilibrium with each
other, and multimerization is essentially an irreversible
reaction.
There is evidence that the aggregation process is specific,
involving certain structural moieties such as helix-helix docking or
cross-ß-sheet formation. Inclusion bodies formed within cells
are relatively pure and do not contain a significant fraction of
other cellular proteins. In addition, mixed aggregation studies of
the P22 coat and tailspike proteins suggest that the tailspike does
not form heterogeneous aggregates with the P22 coat protein. This
methodology of isolating early multimers along the aggregation
pathway has been shown to be applicable to other proteins, such as
the P22 coat protein and carbonic anhydrase II, and may potentially
be used to monitor inclusion body formation or detect amyloid fibil
intermediates associated with Alzheimers disease.
Related Publications:
Margaret A. Speed, Daniel I.C. Wang, and Jonathan
King 1995. "Multimeric
Intermediates in the Pathway to the Aggregated Inclusion Body State
for P22 Tailspike Polypeptide Chains." Protein
Science 4:900-908.
Return to King
Lab Research
Return to King Lab
Homepage
 polymerized by noncovalent association of a monomeric folding
intermediate rather than aggregation of nativelike trimeric species.
These multimers were sensitive to SDS and protease unlike the
thermostable native trimer. Using a modified nondenaturing Western
blot procedure, aggregation intermediates were determined to have
nonnative epitopes in common with productive folding intermediates.
The aggregate epitopes were found on folding and aggregation
intermediates but not on denatured or native tailspike protein. The
nonnative epitope on the folding and aggregation intermediates was
located on the partially folded N-terminus, and this structural
information has implications on the aggregation mechanism.
polymerized by noncovalent association of a monomeric folding
intermediate rather than aggregation of nativelike trimeric species.
These multimers were sensitive to SDS and protease unlike the
thermostable native trimer. Using a modified nondenaturing Western
blot procedure, aggregation intermediates were determined to have
nonnative epitopes in common with productive folding intermediates.
The aggregate epitopes were found on folding and aggregation
intermediates but not on denatured or native tailspike protein. The
nonnative epitope on the folding and aggregation intermediates was
located on the partially folded N-terminus, and this structural
information has implications on the aggregation mechanism.