

The following list of abstracts of publications is ordered chronologically. For a categorical listing of the publications, please navigate to the RESEARCH subsection of this web site.
Click
here to see an article that gives a great overview of protein
folding and misfolding!
Unraveling the Mystery of Protein Folding. W. A. (Bill)
Thomasson (Jonathan King served as science writer). From the
Breakthroughs in Bioscience series from the FASEB
Office of Public Affairs.
The capsid assembly pathways of the dsDNA bacteriophages,
herpesviruses and adenoviruses all proceed through a precursor shell
lacking DNA. These procapsids contain scaffolding proteins required
for assembly but absent from mature virions. The bacteriophage P22
procapsid contains approximately 300 molecules of the 33-kDa gene 8
scaffolding protein, in addition to the 420 molecules of gene 5 coat
protein. During the process of DNA packaging and phage maturation,
all 300 scaffolding molecules are released intact to participate in
subsequent rounds of procapsid assembly. Low concentrations of
guanidine hydrochloride (GuHCl) reproduce the release of scaffolding
from procapsids in vitro, in the absence of DNA. The release was
reversible; when the GuHCl was removed by dialysis, the scaffolding
subunits reentered the extracted capsids to regenerate
morphologically normal procapsids. The subunits presumably exited and
reentered through the channels recently observed at the centers of
the pentamers and hexamers (Prasad, B. V. V., Prevelige, P. E.,
Marietta, E., Chen, R. O., Thomas, D., King, J., and Chiu, W. (1993).
J. Mol. Biol. 231, 65-74). We have utilized this reaction to
investigate the binding of scaffolding subunits within normal
procapsids and to other large structures of coat protein. Procapsids
contained two classes of scaffolding subunits, which may represent
binding of scaffolding to different specific positions within the T =
7 procapsid lattice. These sites became lost or inaccessible upon
phage maturation.
Go to Bacteriophage
P22 Assembly.
The failure of newly synthesized polypeptide chains to reach the
native conformation due to their accumulation as inclusion bodies is
a serious problem in biotechnology. The critical intermediate at the
junction between the productive folding and the inclusion body
pathway has been previously identified for the P22 tailspike
endorhamnosidase. We have been able to trap subsequent early
intermediates in the in vitro pathway to the aggregated inclusion
body state. Nondenaturing gel electrophoresis identified a sequential
series of multimeric intermediates in the aggregation pathway. These
represent discrete species formed from noncovalent association of
partially folded intermediates rather than aggregation of native-like
trimeric species. Monomer, dimer, trimer, tetramer, pentamer, and
hexamer states of the partially folded species were populated in the
initial stages of the aggregation reaction. This methodology of
isolating early multimers along the aggregation pathway was
applicable to other proteins, such as the P22 coat protein and
carbonic anhydrase II.
Go to Aggregation
Intermediates.
An unexpected aspect of the expression of cloned genes is the
frequent failure of newly synthesized polypeptide chains to reach
their native state, accumulating instead as insoluble inclusion
bodies. Amyloid deposits represent a related state associated with a
variety of human diseases. The critical folding intermediates at the
juncture of productive folding and the off pathway aggregation
reaction have been identified for the phage P22 tailspike and coat
proteins. Though the parallel beta coil tailspike is thermostable, an
early intracellular folding intermediate is thermolabile. As the
temperature of intracellular folding is increased, this species
partitions to inclusion bodies, a kinetic trap within the cell. The
earliest intermediates along the in vitro aggregation pathway,
sequential multimers of the thermolabile folding intermediates, have
been directly identified by native gel electrophoresis. Temperature
sensitive folding (tsf) mutations identify sites in the beta coil
domain which direct the junctional intermediate down the productive
pathway. Global suppressors of tsf mutants inhibit the pathway to
inclusion bodies, rescuing the mutant chains. These mutants identify
sites important for avoiding aggregation. Coat folding intermediates
also partition to inclusion bodies as temperature is increased. Coat
tsf mutants are suppressed by overexpression of the GroE chaperonin,
indicating that the thermolabile intermediate is a physiological
substrate for GroE. We suggest that many proteins are likely to have
thermolabile intermediates in their intracellular folding pathways,
which will be precursors to inclusion body formation at elevated
temperatures, and therefore substrates for heat shock
chaperonins.
Assembly of the icosahedral shells of the dsDNA bacteriophages,
herpesviruses and adenoviruses all require proteins not found in the
mature virion, termed scaffolding proteins. The bacteriophage P22
precursor procapsid contains approximately 300 scaffolding molecules
within a shell composed of 420 coat protein subunits. Though nonsense
mutants are common, few mutants affecting the functions of the
scaffolding protein have been recovered. We report here the isolation
and characterization of new missense mutants unable to form
infectious virions under restrictive conditions. These mutant
scaffolding subunits were competent for protein folding and capsid
assembly under restrictive conditions. Two mutants were defective in
assembly into the procapsid of the portal complex, which serves as
the channel through which DNA is packaged. These mutations may
identify a region of the scaffolding protein required for interaction
with the portal subunits. Two mutants in a different region of the
sequence were impaired in scaffolding release from the procapsid both
in vivo and in vitro. These mutations may identify a new domain
required for scaffolding release. Scaffolding release appeared to be
required for capsid expansion; in turn, scaffolding release seemed to
depend upon the presence of a portal. This may help to order the
pathway of events in phage maturation.
Go to Bacteriophage
P22 Assembly.
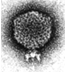
The procapsids of bacterial viruses are the products of the
polymerization of coat and scaffolding subunits, as well as the
precursors in DNA packaging. Electron cryo-microscopy has been used
to study the three-dimensional structures of bacteriophage P22
procapsids containing wild-type and mutant scaffolding proteins. The
scaffolding mutant structure has been resolved to 19 ™
resolution and agrees with the 22 ™ resolutionwild-type
procapsid reconstruction. Both procapsid reconstructions contain an
outer icosahedral coat protein shell and an inner scaffolding protein
core. The outer coat protein forms a T = 7 icosahedral lattice of
pentons and skewed hexons. The distinctive channels at the centers of
the pentons and hexons, first observed in the lower resolution empty
procapsid structure (Prasad et al., 1993), are present. Computational
isolation of the skewed hexon shows the presence of a local two-fold
axis which reduces the number of unique conformations in the
asymmetric unit to four at this resolution. The four unique subunits
have been classified into three distinct classes based upon the shape
of the upper domain, and the presence of a channel leading to the
inner coat protein surface. In addition, at the inner surface of the
coat protein, finger-like regions which extend towards the
scaffolding protein core are present in two of the subunits. The
finger-like regions suggest the presence of an ordered interaction
between the inner coat protein and the scaffolding protein. However,
an icosahedral scaffolding protein shell is not formed, and the inner
most scaffolding protein core is not packed with icosahedral
symmetry.
Go to Bacteriophage
P22 Assembly.
 The Jonathan King Lab Homepage
The Jonathan King Lab Homepage