|
BioSyM Research
Thrust 3: In vitro cellular systems engineering
|
|
|
|
|
3D Microfluidic Platform for cancer drug screening |
Hypoxia on a Chip |
Induction of Angiogenesis by PHis |
Sprout Formation in Angiogenesis |
In thrust 3, we develop platforms and methods to quantify behavior of cells within mixed, structurally complex populations and systems.C ell culture has historically been conducted in tissue culture dishes with a single cell type. However, new methods now allow cells to be grown in vitro within a more physiologically representative 3-dimensional (3D) microenvironment, in co-culture with other cell types, and under progressively greater control of the numerous factors that influence cell behavior in vitro, temporal and spatial gradients of cytokines, growth factors, and oxygen tensions, to name just a few. Such devices, methods, and associated computational analysis of timelapsed responses can aid in creating in vitro assays that more accurately mimic conditions in vivo. We use these in vitro platforms to study drug screening for drugs to prevent the first steps in metastatic cancer, models of angiogenesis, systems to test interactions between fibroblasts and endothelial cells in the creation of new vasculature, and studies of cell migration and interaction between various pluripotent cells and excised tissue specimens.spatial gradients of cytokines, growth factors, and oxygen tensions, to name just a few. Such devices, methods, and associated computational analysis of timelapsed responses can aid in creating in vitro assays that more accurately mimic conditions in vivo.
Capabilities developed:
- Design and fabrication of microfluidic devices for 3D cell culture: Various designs to suit cell types and study target (drug screening, angiogenesis, cell encapsulation, electro-mechanical stimulation).
- Protocols for 3D cell culture in microfludic devices.
- Computational models using finite volume and finite element analysis to predict cell migration.
- A suite of image processing software which combines heterogeneous images, acquired with diverse imaging modalities to optimally interpret data. Imaging modalities include
- transmitted light imaging such as bright field, dark field, phase-contrast, and Differential Interference Contrast (DIC) microscopy,
- epi-fluorescence and confocal microscopy.
People
|
Research Highlights
|
|
1. Development of drug screening platform for therapeutic EMT blocking agents in a 3D microenvironment
Over 90% of all cancer-related deaths are consequences of metastatic tumors. Epithelial–Mesenchymal Transition (EMT) plays a critical role in the early stages of dissemination of carcinoma leading to metastatic tumors. Current therapeutic regimens, however, have been ineffective in the cure of metastatic cancer, thus an urgent need exists to revisit existing protocols and to improve the efficacy of newly developed therapeutics. Strategies based on preventing EMT could potentially contribute to improving the outcome of advanced stage cancers. To achieve this goal new assays are needed to identify targeted drugs capable of interfering with EMT or to revert the mesenchymal-like phenotype of carcinoma to an epithelial-like state. Current assays are limited to examining the dispersion of carcinoma cells in isolation in conventional 2-dimensional (2D) microwell systems, an approach that fails to account for the 3-dimensional (3D) environment of the tumor or the essential interactions that occur with other nearby cell types in the tumor microenvironment. Here we present a microfluidic system that integrates tumor cell spheroids in a 3D hydrogel scaffold, in close co-culture with an endothelial monolayer. Drug candidates inhibiting receptor activation or signal transduction pathways implicated in EMT have been tested using dispersion of A549 lung adenocarcinoma cell spheroids as a metric of effectiveness. We observe significant differences in response to drugs between 2D and 3D, and between monoculture and co-culture.
Two different cancer cell lines, A549 (lung) and T24 (bladder), were tested. Dose-response assays of Src inhibitor AZD-0530 were obtained for relative dispersion as a metric of epithelial-mesenchymal transition (EMT). Although dispersion of A549 cells could be inhibited, none of the doses of AZD-0530 inhibited T24 dispersion. Hepatocyte growth factor (HGF) signaling was further identified that plays a role in T24 dispersion. This 3D microfluidic co-culture platform provides an in vivo-like surrogate for anti-invasive and anti-metastatic drug screening. It will be particularly useful for defining combination therapy for aggressive tumors such as invasive bladder carcinoma.
|
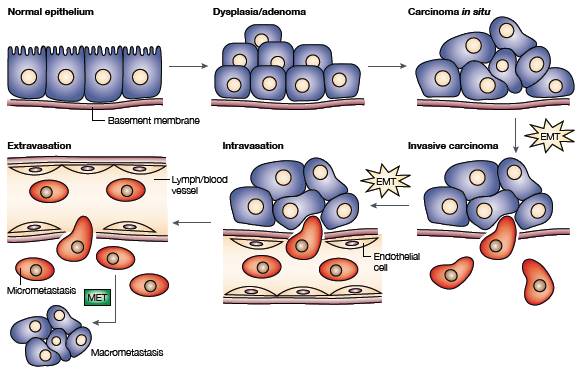
Scheme of the EMT (Epithelial–Mesenchymal Transition) and Circulating Tumour Cell extravasation (Thiery, J. P. Epithelial–mesenchymal transitions in tumour progression, Nature Publishing Group 2, 442–454 (2002). |
|
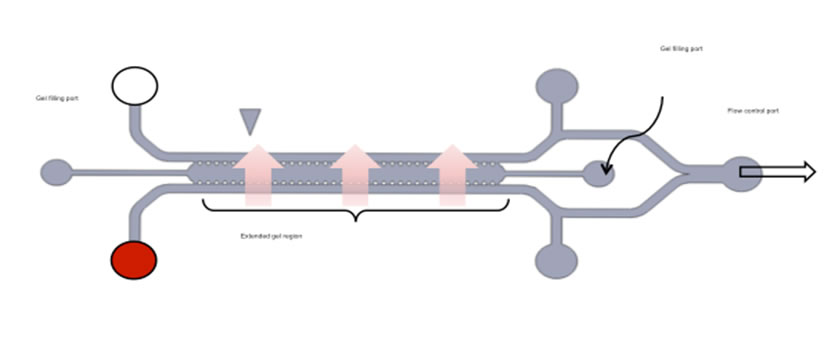
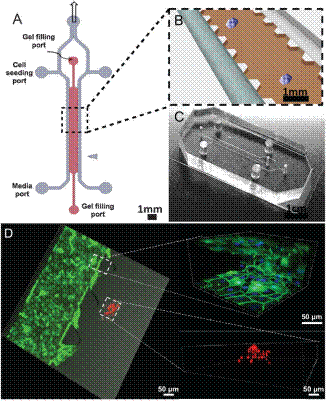
Schematic and photograph of a 3D co-culture microfluidic device.(A) Schematic diagram of device layout depicts the inlets for injecting cells, filling collagen, and replenishing medium. (B) Enlarged view of gel region and the HUVEC-lined channel. Cytokines in conditioned medium from the HUVEC monolayer diffuse into the gel region triggering spheroids to undergo EMT. (C) Photograph of the PDMS-molded device bonded on a glass cover-slip. (D) A 3D co-culture image combining phase contrast and fluorescence with enlarged HUVECs monolayer structure and 3D cancer spheroid dispersion. Blue: Hoechst; green: VE-Cadherin; red: nuclei mCherry. |
|
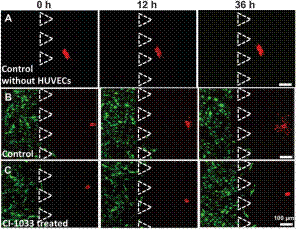
Fluorescent images in time-series showing A549 cell dissemination in the 3D collagen gel. (A) Control condition in the absence of a HUVEC monolayer, i.e., 3D monoculture. (B) Control condition in the presence of a HUVEC monolayer in the side channel, i.e. 3D co-culture. (C) EGF-targeted drug (300 nM of CI-1033) applied in the presence of a HUVEC monolayer. Red: nuclei of A549 cells; green:HUVEC. Triangles show the PDMS posts on the edge of collagen gel. |
2. Induction of Angiogenesis in Microfluidic Devices using Prolyl Hydroxylase Inhibitors and Sphingosine-1 Phosphate
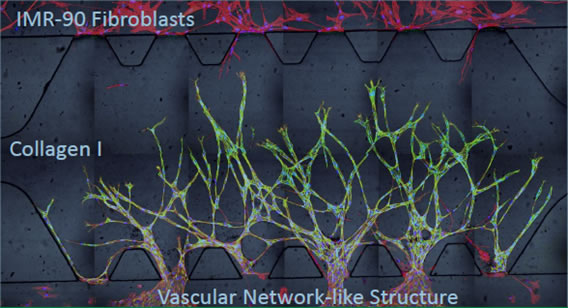
The use of microfluidic devices to study angiogenesis, the sprouting of new blood vessels from pre-existing vessels, in 3D has been widely adopted for the creation of in vitro assays that are closer to in vivo conditions. Our recent work develops microfluidic devices to study synergistic effects of two different angiogenic compounds, prolyl hydroxylase inhibitors (PHis) and sphingosine 1-phosphate (S1P) in the presence of fibroblasts. PHis are compounds that prevent the degradation of hypoxia inducible factor-1α (HIF-1α) in vivo thus upregulating various angiogenic factors including VEGF as a response to the induced pseudo-hypoxic condition under normoxia. S1P, the other hand, is a lysophospholipid that mediates multiple biological events including angiogenesis through specific cell surface G-protein coupled receptors. The synergistic effects of PHis and S1P are studied in an in vitro microfludic platform where endothelial cells are co-cultured with alginate bead encapsulated-fibroblasts and subjected to the stimulation of ciclopirox olamine (CPX, a type of PHis) and S1P. We find that the interactions between endothelial cells and fibroblasts are essential in maintaining immature sprouts and converting them into functional vessels with lumens and these effects are most prominent when CPX works synergistically with S1P.
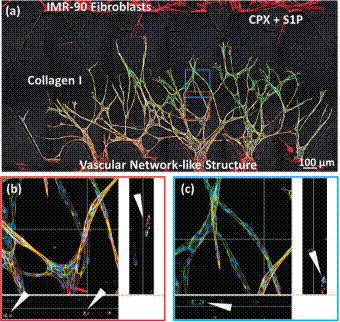
|
In combination, CPX (PHi) and S1P induced vascular network structure with defined lumina. (a) Stitched; (b) and (c) sectioned 20X confocal images of Hoechst stained nuclei (blue), rhodamine phalloidin stained actin (red) and Alexa fluor 488 immunostained VE-cadherin (green) show sprouts that anastomose under 8 mM CPX and 250 nM S1P induction. White arrows indicate lumina of endothelial sprouts with diameters around 30 mm. Scale bar = 100 mm. |
We demonstrated that a cross gradient of mutually stimulating factors was generated as each cell type responded differently to the compounds, especially to the PHi. EC showed increased secretion of placental growth factor, endothelin-1, epidermal growth factor and interleukin-8 while fibroblasts showed upregulated secretion of hepatocyte growth factor, insulin-like growth factor-binding protein 2, urokinase plasminogen activator and most importantly, vascular endothelial growth factor (VEGF). We also showed that S1P induced migration was S1P receptor 1 (S1P1) dependent and more importantly the expression of S1P1 on EC could be modulated by both PHi and S1P. The downregulation of either VEGF or S1P1 showed attenuated capillary sprouting and also change of sprouting morphologies. The proangiogenic monocyte chemotactic protein (MCP)-1 was mainly secreted by fibroblasts, but only in the presence of EC-conditioned medium (CM). Antibody interference with MCP-1 also reduced the sprouting response. The EC-fibroblasts interactions could be replaced by neither fibroblasts CM nor EC-cancer cells crosstalk. These observations not only confirm the importance of EC-fibroblasts crosstalk in angiogenesis but also suggest that the combination of PHi and S1P enhances this synergy and thus might be of therapeutic value for tissue repair and regeneration.
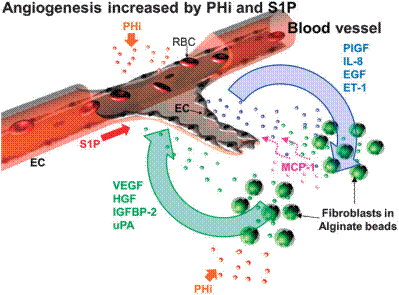
|
Schematic diagram of PHi + S1P-driven capillary sprouting. PHi exerts differential effects on EC and fibroblasts that lead to upregulation of PlGF, IL-8, EGF and ET-1 from EC and upregulation of VEGF, HGF, IGFBP-2 and uPA from fibroblasts. The soluble factors secreted by fibroblasts and EC together with S1P promote angiogenesis. Fibroblast-derived MCP-1 is induced by endothelial factors and it also supports the growth of capillaries. |
3. Oxygen Levels in Thermoplastic Microfluidic Devices during Cell Culture
We developed a computational model to predict oxygen levels in microfluidic plastic devices during cell culture. This model is based on experimental evaluation of oxygen levels. Conditions are determined that provide adequate oxygen supply to two cell types, hepatocytes (HEP) and endothelial cells (EC), either by diffusion through the plastic device, or by supplying a low flow rate of medium. PDMS devices where fabricated with soft lithography techniques. Alternatively, the COC (cyclic olefin copolymer) and PMP (poly-methyl pentene) chips were injection molded. Oxygen-sensitive foils (Visisens, Germany) were bonded to the bottom of the devices. All oxygen measurements were evaluated using the Visisens software after calibration against two reference solutions.
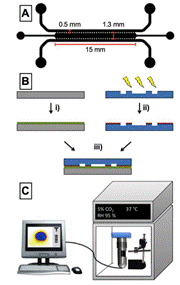
|
(A) Schematic of the microfluidic chip design and dimensions. (B) Device assembly: i) oxygen sensor foil is spin- coated with PDMS. ii) plasma-activated thermoplastic is exposed to APTS solution. iii) plasma-bonding of substrate and microfluidic chip. (C) Experimental setup: the microfluidic chip is positioned on top of the handheld microscope, which is placed in the incubator for cell culture. The microscope is connected to a laptop for data collection and analysis. |
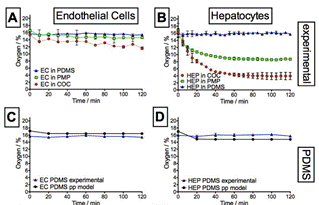
|
Summary of experimental (A+B) and simulated oxygen levels (C-H) for endothelial and hepatocytes culture in microfluidic devices made from PDMS (C+D), PMP (E+F) and COC (G+H). Coloured symbols represent experimental data, which are compared to simulation results (black symbols).
HEP depleted almost all oxygen in the impermeable COC device, reaching 4% within 60 min (B). The mildly hypoxic conditions observed in PMP devices (<11%) are very similar to oxygen levels prevailing in vivo. In thin PDMS devices, however, oxygen is readily replenished from the environment, resulting in stabilization of oxygen levels at ~16% for both HEP and EC culture (A+B).
Cell culture in PMP devices gave rise to intermediate oxygen levels in all cases due to its intrinsic properties (high oxygen permeability compared to COC), making PMP a promising candidate for commercial production of microfluidic cell culture devices. |
4. Concave Microwells for the Formation of 3D Multicellular Cancer Aggregates for Drug Screening
3D tissue-like cultures are now widely used as in vitro models of in vivo cellular functions such as embryonic development, wound healing, and malignancy. Compared with a 2D monolayer culture, a 3D model better mimics the in vivo tissue like cellular features in morphology, cell–cell interaction, signal transduction, and mechanical stimulation, thus providing a physiologically and pathologically relevant environment. Multicellular 3D aggregates have proven useful as a model for directing specifi c cell lineages using embryonic stem cell-formed embryoid bodies, and studying collective cell migration in cancer.
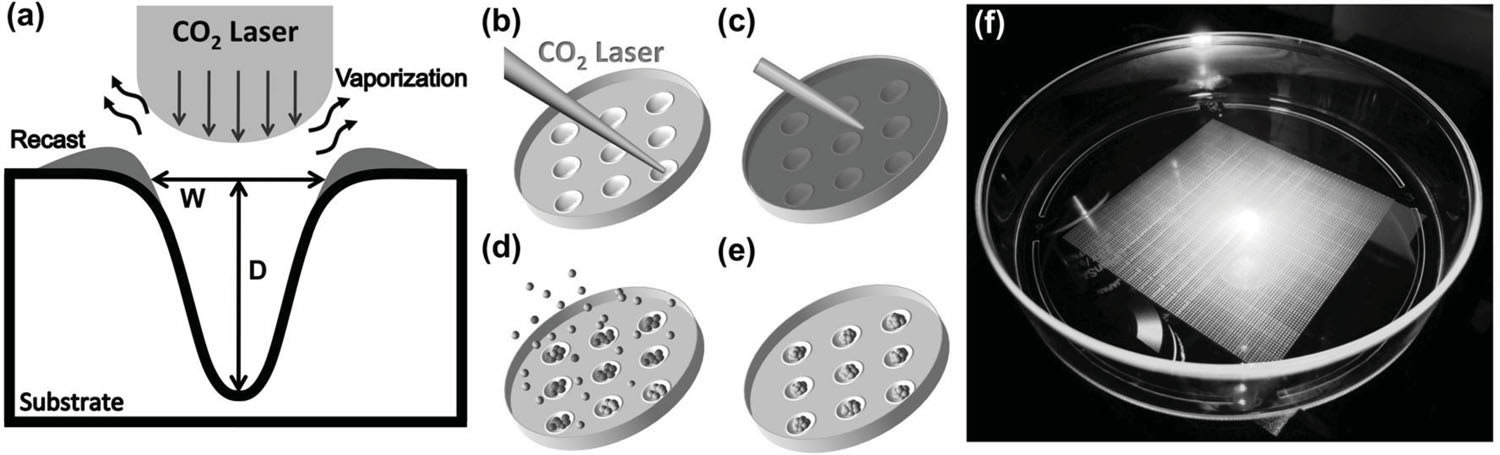
Microwell technology has revolutionized many aspects of in vitro cellular studies from 2D traditional cultures to 3D in vivo-like functional assays. However, existing lithography-based approaches are often costly and time consuming. This study presents a rapid, low-cost prototyping method of CO2 laser ablation of a conventional untreated culture dish to create concave microwells used for generating multicellular aggregates, which can be readily available for general laboratories. Polymethylmethacrylate (PMMA), polydimethylsiloxane (PDMS), and polystyrene (PS) microwells are investigated, and each produces distinctive microwell features. Among these three
materials, PS cell culture dishes produce the optimal surface smoothness and roundness. A549 lung cancer cells are grown to form cancer aggregates of controllable size from ≈40 to ≈80 μ m in PS microwells. Functional assays of spheroids are performed to study migration on 2D substrates and in 3D hydrogel conditions as a step towards recapitulating the dissemination of cancer cells. Preclinical anti-cancer drug screening is investigated and reveals considerable differences between 2D and 3D conditions, indicating the importance of assay type as well as the utility of the present approach.
|
Schematic diagram of the microwell fabrication process for generation of 3D aggregates. a) Illustration of laser ablation process. Substrate is melted by CO 2 laser energy distributed in a Gaussian profi le. The ablation leaves a concave well with a recast zone at its edge. b) Laser drilling via an XY mobile focus carriage traveling to the location of interest. c) A thin layer of pluronic was deposited to prevent cell attachment on the substrate. d) Cell seeding. e) Formation of aggregates after 4–5 d. f) A snapshot of microwell array pattern in a cell culture dish. |
|
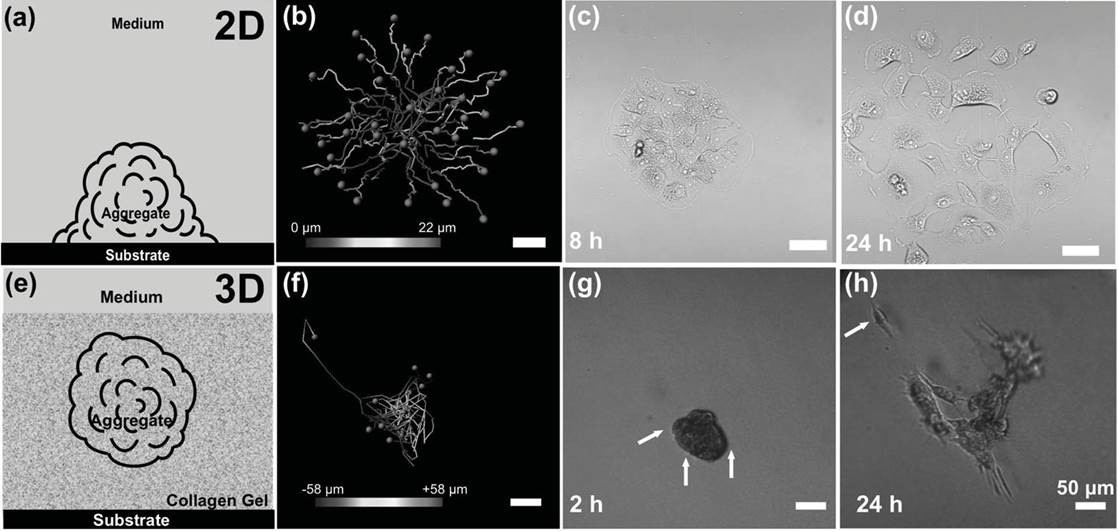
Schematic diagram and cell migration path over 24 h in 2D and 3D conditions. a) Diagram of MCAs in direct contact with the substrate, where dispersion occurs on the 2D surface. b) Single cells tracking over 24 h in 2D conditions. c,d) Phase-contrast image of an MCA spreading at 8 and 24 h in 2D conditions. e) Diagram of MCAS in 3D suspension in native type I collagen. f) Cell tracking over 24 h under 3D conditions. g) Phase-contrast
image of an MCA embedded in 3D collagen matrix showing early formation of pseudopodia at 2 h, indicated by arrows. h) The same MCA broke up
after 24 h, and a single escaped cell was observed (indicated by the arrow). Color bars represent the position along the Z direction. |
5. Tracking of angiogenic sprouting cells interacting with the gel matrix using a Bayesian filtering approach
Cells in the human body migrate to perform numerous biological functions, including development and wound healing. For years, cell migration has been studied with two-dimensional (2D) on-the-gel assays using 2D imaging and cell tracking techniques. When cells migrate in three-dimensional (3D) environments, they exhibit strikingly different behaviors. In 3D, cells migrate by remodeling and cleaving the surrounding gel whereas in 2D, they simply move across a free surface. However, their migration behavior and interactions with the surrounding gel are still poorly understood. To help understand 3D migration and cell-gel interactions, we aim to develop a new automated image processing methodology which will provide useful insights into how a vascular structure is formed as a collection of migratory cells. We present a new approach to incorporating information from heterogeneous images of migrating cells in 3D gel. We study 3D angiogenic sprouting, where cells burrow into the gel matrix, communicate with other cells and create vascular networks. We combine time-lapse fluorescent images of stained cell nuclei and transmitted light images of the background gel to track cell trajectories. The nuclei images are sampled less frequently due to photo toxicity. Hence, 3D cell tracking can be performed more reliably when 2D sprout profiles, extracted from gel matrix images, are effectively incorporated. We employ a Bayesian filtering approach to optimally combine the two heterogeneous images with different sampling rates. We construct stochastic models to predict cell locations and sprout profiles and condition the likelihood of nuclei location by the sprout profile. The conditional distribution is non-Gaussian and the cell dynamics is non-linear. To jointly update cell and sprout estimates, we use a Rao–Blackwell particle filter. Simulation and experimental results show accurate tracking of multiple cells along with sprout formation, demonstrating synergistic effects of incorporating the two types of images.
|
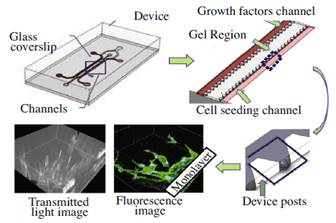
Experiments are carried out in a microfluidic flow chamber with 37 migration regions. We inject collagen gel between the two channels. Cells are seeded in one channel and growth factors are introduced in the other. A confocal microscope captures fluorescence and transmitted light images of each migration region, including the cell monolayer. |
|
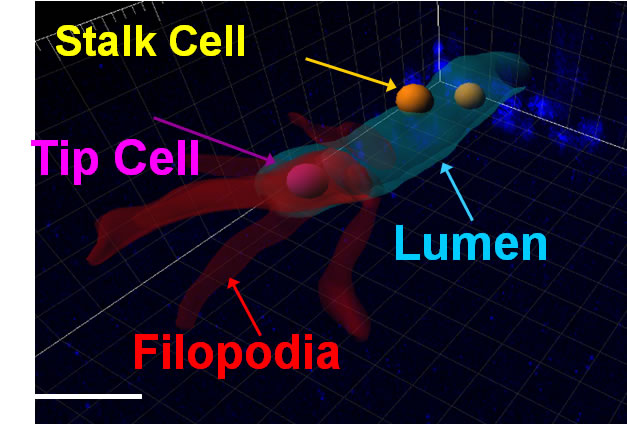
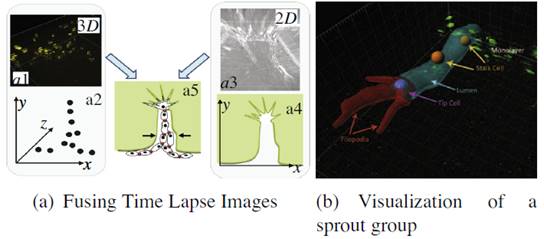
Incorporating 3D with 2D information. (a) 3D cell nuclei centroid ocations (a2) are obtained from the fluorescent images (a1). The bright field images (a3) provide 2D information about the sprout (a4). The goal is to combine this information (a5). In the sprout group visualized in (b), we visualize a lumen (cyan tube), created by a tip cell (purple sphere). Tip cells are characterized by extensive filopodia, indicated in red. They are followed by stalk cells (orange spheres), which migrate within the lumen. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) |
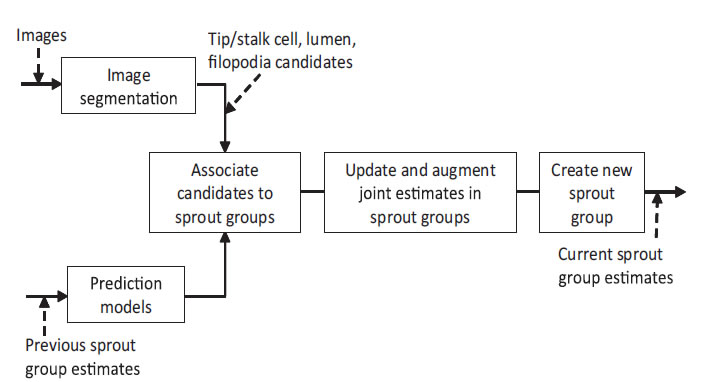
Overview of the tracking system. Multiple sprouts, are treated as separate groups of cells, lumen and filopodia. We predict these parameters using motion models. The segmented nuclei, lumen and filopodia candidates are associated firstly with a particular sprout, and then with existing tracks in its joint estimate. Matched candidates are updated to an existing track. New sprouts may be created for dissociated sprout profiles. New nuclei, lumen and filopodia tracks in a sprout may be augmented to a joint estimate. Tracks and sprouts may also be terminated.
6. Quantification of Magnetically Induced Changes in Extra Cellular Matrix (ECM) Local Apparent Stiffness
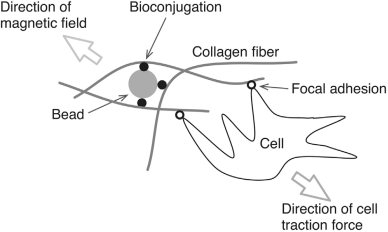
The mechanical properties of the materials surrounding cells influence the behaviour of cells in differentiation, proliferation, and apoptosis. We have developed a new way to change one mechanical property of the extracellular matrix (ECM), namely the stiffness, via embedded magnetic beads. These beads are conjugated in an ECM via streptavidin-coated beads that bind to biotinylated collagen fibres, then applying an external magnetic field to exert forces on the beads in order to manipulate the local strain and then the effective stiffness of the ECM. The change in the micro and macro Young’s elastic modulus of the ECM sample, when switching on or off the magnetic field, was determined by atomic force microscopy-enabled indentation (developed in Thrust 2) and uniaxial compression, respectively. The experimental results demonstrated that, by embedding magnetic beads in an ECM through bio-conjugation between the streptavidin-coated beads and the collagen fibers, the stiffness of the ECM can be actively manipulated by the application of an external magnetic field. A separate study was done by allowing endothelial cells (ECs) to migrate though the magnetic bead embedded ECM ensemble with magnetic field and without magnetic field. A VEGF gradient was also added to promote the sprouting of ECs. The results from this work have demonstrated the possibility of creating desired stiffness gradients in an ECM in vitro to influence endothelial cell behaviour. Electromagnetic needles (EMN) were designed and fabricated to be installed in the existing confocal microscope for the manipulations of magnetics beads during angiogenesis. These EMNs are designed such that their magnetic fields are focused on a smaller region to allow single magnetic bead manipulations.
|
Schematic illustration of bead-embedded ECM and interaction between collagen fibers and cells via focus adhesions under the influence of an external magnetic field. This scenario provides motivation for the work reported in this article on the determination of the change in the apparent local stiffness of the ECM under the influence of an external magnetic field. |
7. An Integrated Cell Migration Model Incorporating Cytoskeleton Remodeling, Focal Adhesion Dynamics, and Actin Motor Activity
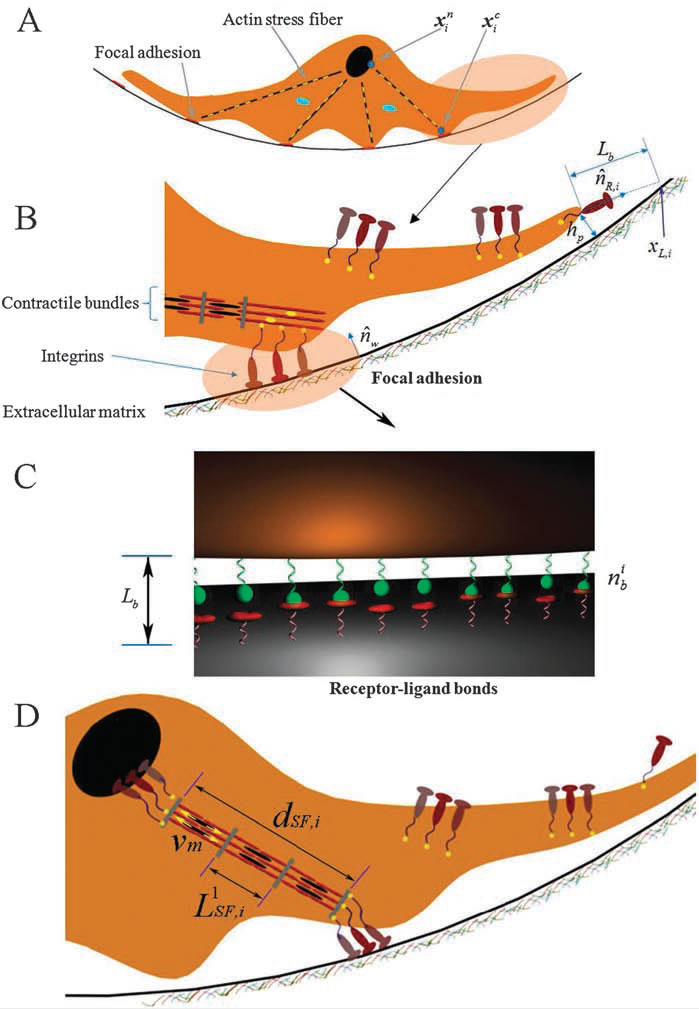
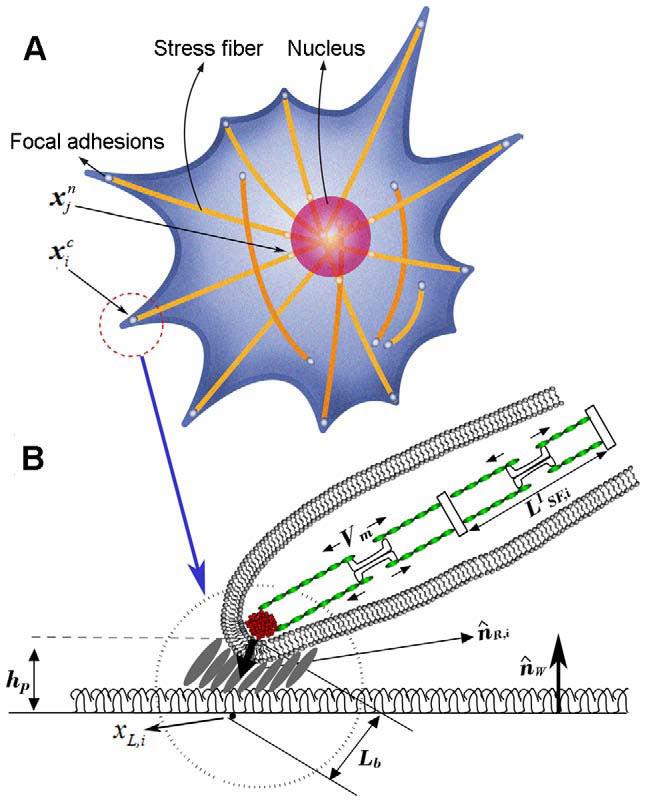
An integrative cell migration model incorporating focal adhesion (FA) dynamics, cytoskeleton and nucleus remodeling and actin motor activity is developed for predicting cell migration behaviors on 3-dimensional curved surfaces, such as cylindrical lumens in the 3-D extracellular matrix (ECM). The work is motivated by 3-D microfluidic migration experiments suggesting that the migration speed and direction may vary depending on the cross sectional shape of the lumen along which the cell migrates. In this paper, the mechanical structure of the cell is modeled as double elastic membranes of cell and nucleus. The two elastic membranes are connected by stress fibers, which are extended from focal adhesions on the cell surface to the nuclear membrane. The cell deforms and gains traction as transmembrane integrins distributed over the outer cell membrane bind to ligands on the ECM, form focal adhesions, and activate stress fibers. Probabilities at which integrin ligand–receptor bonds are formed as well as ruptures are affected by the surface geometry, resulting in diverse migration behaviors that depend on the curvature of the surface. Monte Carlo simulations of the integrative model reveal that (a) the cell migration speed is dependent on the cross sectional area of the lumen with a maximum speed at a particular diameter or width, (b) as the lumen diameter increases, the cell tends to spread and migrate around the circumference of the lumen, while it moves in the longitudinal direction as the lumen diameter narrows, (c) once the cell moves in one direction, it tends to stay migrating in the same direction despite the stochastic nature of migration. The relationship between the cell migration speed and the lumen width agrees with microfluidic experimental data for cancer cell migration.
To understand three-dimensional crawling behaviour of a single stalk cell on the surface of the angiogenic lumen, we developed an integrated cell migration model incorporating three key mechanisms of cell biology, consisting of remodelling of the cell and nuclear membranes, focal adhesion dynamics, and actin motor activity. After we successfully compared our model with an existing experimental work of spontaneous cancer cell migration in a confined microfluidic device, we predicted stalk cell migration in various circular tubes. When the cell migrates on the wall of a wide lumen, the cell tends to stretch out along the circumference, where the radius of curvature is smaller than that of the longitudinal direction, resulting in a high probability for transverse migration. The new cell migration model can be further developed toward a more complex model with the inclusion of cell–cell interactions to predict emergent behaviours of collective cell migrations in various geometries. |
|
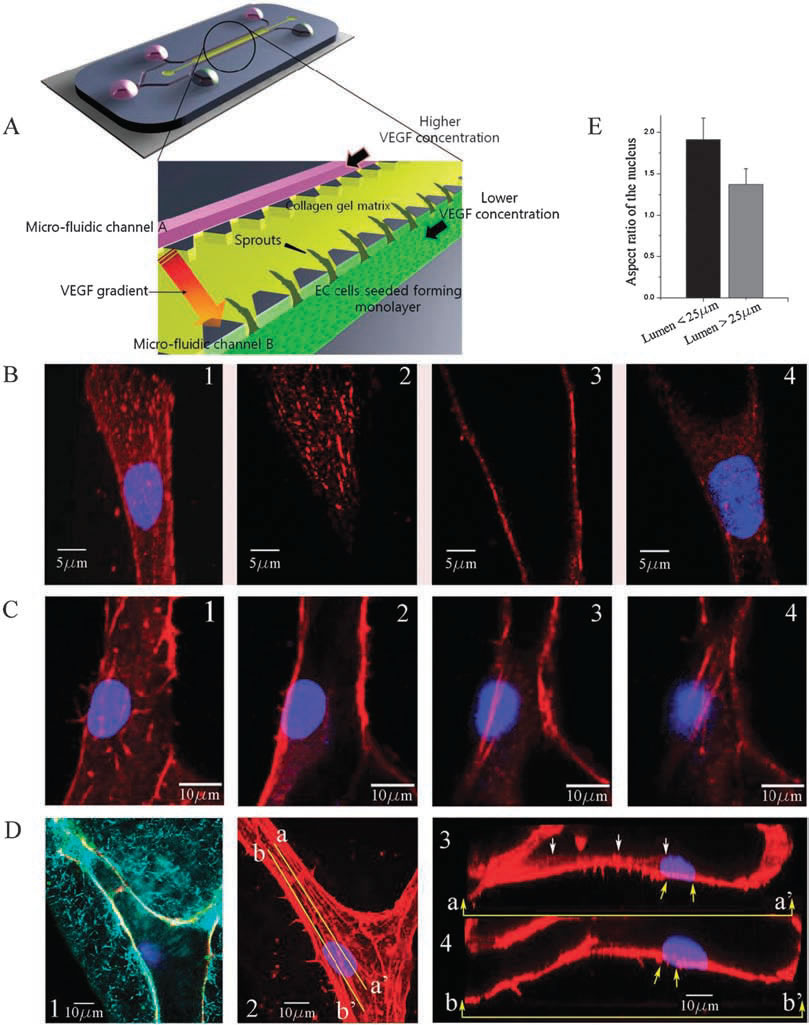
Experimental observations of focal adhesion sites and actin stress fibers on the lumen in a 3-D collagen matrix: (A) 3-D microfluidic assay with hMVECs seeded on one side of the collagen gel. Higher concentration of VEGF is supplied to channel A and lower concentration of VEGF to channel B so that a gradient of VGEF is created across the gel. (B) Stalk cells migrating into the gel are observed; (1) collapsed confocal 3-D image (120X) showing a stalk cell migration along a narrow lumen, and slices at selected heights of (2) z=6.3 um (top), (3) 10.08 um (middle), and (4) 16.38 um (bottom); nucleus and focal adhesion sites are stained with Hoechst (blue) and vinculin (red), respectively. (C) Actin stress fibers in a larger lumen with a magnification of 120X; sectional slices showing stress fibers at selected heights; (1) z = 0 um, (B) 0.76 um, (3) 5.32 um and (4) 6.84 um; nucleus and actin stress fibers are stained with Hoechst (blue) and Rhodamine phalloidin (red), respectively. 6.84 um; nucleus and actin stress fibers are stained with Hoechst (blue) and Rhodamine phalloidin (red), respectively. (D(1)) A reflectance microscopy image showing the creation of a hole by a migrating tip cell in the 3-D collagen matrix; nucleus and vascular endothelial (VE)-cadherins are stained with Hoechst (blue) and anti-VE cadherin (green), (D(2)) collapsed confocal 3D image (120X); nucleus and actin stress fibers
are stained with Hoechst (blue) and Rhodamine phalloidin (red), respectively, (D(3)) the longitudinal cross sectional view across line aa' shown in
D(2); short actin stress fibers are seen beneath the nucleus, (D(4)) the longitudinal cross sectional view across line bb' shown in D(2); long stress
fibers (yellow arrows) connected to the nucleus extend towards the leading edge of the migrating cell. (E) Quantification of the aspect ratio of the
nucleus under two conditions when the cell migrates into the lumen with a diameter less than 25 mm and larger than 25 mm. Data are means +/- SD.
In each case 35 cells were analysed. |
|
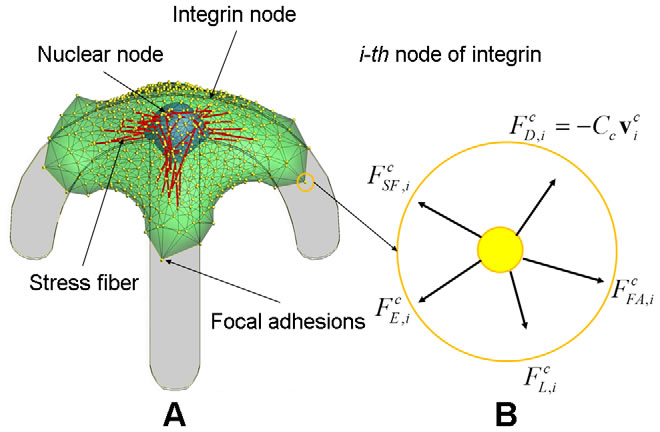
3-D integrated cell migration model: (A) 2-D schematic representation of a cell migration model on the curved ubstrate, showing deformable cell and nuclear membranes, and actin stress fibers, (B) a magnified view in A showing the structure of focal adhesion including the attachment of the end of stress fibers through an integrin node to the underlying extracellular matrix, (C) a magnified view in B illustrating a stochastic ligand–receptor bonding process at the focal adhesion site, and (D) a magnified view in A showing the structure of actin stress fibers connected to a nucleus node. |
8. Dynamic Modeling of Cell Migration and Spreading Behaviors
An integrative cell migration model incorporating focal adhesion (FA) dynamics, cytoskeleton and nucleus remodeling, actin motor activity, and lamellipodia protrusion is developed for predicting cell spreading and migration behaviors. This work is motivated by two experimental works: (1) cell migration on 2-D substrates under various fibronectin concentrations and (2) cell spreading on 2-D micropatterned geometries. These works suggest (1) cell migration speed takes a maximum at a particular ligand density (,1140 molecules/mm2) and (2) that strong traction forces at the corners of the patterns may exist due to combined effects exerted by actin stress fibers (SFs). The integrative model of this paper successfully reproduced these experimental results and indicates the mechanism of cell migration and spreading. In this paper, the mechanical structure of the cell is modeled as having two elastic membranes: an outer cell membrane and an inner nuclear membrane. The two elastic membranes are connected by SFs, which are extended from focal adhesions on the cortical surface to the nuclear membrane. In addition, the model also includes ventral SFs bridging two focal adhesions on the cell surface. The cell deforms and gains traction as transmembrane integrins distributed over the outer cell membrane bond to ligands on the ECM surface, activate SFs, and form focal adhesions. The relationship between the cell migration speed and fibronectin concentration agrees with existing experimental data for Chinese hamster ovary (CHO) cell migrations on fibronectin coated surfaces. In addition, the integrated model is validated by showing persistent high stress concentrations at sharp geometrically patterned edges. This model will be used as a predictive model to assist in design and data processing of upcoming microfluidic cell migration assays.
 |
 |
| Dynamic model of cell migration. A) Integrated cell migration model consisting of the cytoskeleton, the nucleus, N integrin nodes on the surface of cytoskeleton, N nuclear nodes on the surface of nucleus, and two types of actin SFs which connect the integrin node to the nuclear node and between integrin nodes; a top view of the model showing triangular mesh network of double membranes of cytoskeleton and nucleus. B) the free body diagram of the i-th integrin node in the circle marked in A) where five external forces are acting. Note that, while shown in 2-D, the force balance exists in 3-D. |
|
3-D integrated cell migration model A) schematic representation of cell migration model on the planar substrate, showing deformable cell and nuclear membranes, focal adhesions, and actin SFs, B) a magnified view in A) showing the structure of focal adhesion including the attachment of the end of SFs through an integrin node to the underlying extracellular matrix, illustrating a stochastic ligand-receptor bonding process at the focal adhesion site, and showing the structure of actin SFs. Note that, A) and B) represent top and side views, respectively. |
9. Dll4-containing exosomes induce capillary sprout retraction in a 3D microenvironment
Delta-like 4 (Dll4), a membrane-bound Notch ligand, plays a fundamental role in vascular development and angiogenesis. Dll4 is highly expressed in capillary endothelial tip cells and is involved in suppressing neighboring stalk cells to become tip cells during angiogenesis. Dll4-Notch signaling is mediated either by direct cell-cell contact or by Dll4-containing exosomes from a distance. However, whether Dll4-containing exosomes influence tip cells of existing capillaries is unknown. Using a 3D microfluidic device and time-lapse confocal microscopy, we show here for the first time that Dll4-containing exosomes causes tip cells to lose their filopodia and trigger capillary sprout retraction in collagen matrix. We demonstrate that Dll4 exosomes can freely travel through 3D collagen matrix and transfer Dll4 protein to distant tip cells.
Upon reaching endothelial sprout, it causes filopodia and tip cell retraction. Continuous application of Dll4 exosomes from a distance lead to significant reduction of sprout formation. This effect correlates with Notch signaling activation upon Dll4-containing exosome interaction with recipient endothelial cells. Furthermore, we show that Dll4-containing exosomes increase endothelial cell motility while suppressing their proliferation. These data revealed novel functions of Dll4 in angiogenesis through exosomes.
|
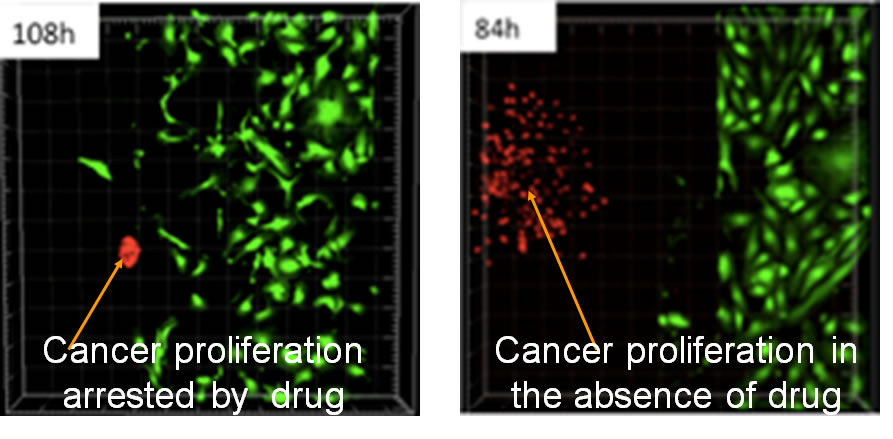
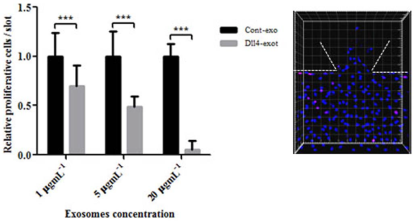
Dll4 exosomes reduced cell proliferation when applied in the gradient. HMVECs were seeded into MFDs after formation of the monolayer they were starved and conditioned with VEGF for three days for sprouts to form. On day three, exosomes were applied in the gradient and incubated for 24 hours. They were fixed and stained on the following day with Ki67 antibody and imaged with the confocal microscope. The total and Ki67 positive cells both in the monolayer and the gel region were counted using Imaris software. The statistical analysis were carried out using Prism software. The experiment was performed with three concentrations of exosomes and three independent repeats. Dll4 exosomessignificantly reduced cell proliferation at higher concentrations *** P , 0.01 but had no significant effect with 5 mgmL21 p 5 0.0537 and 1 mgmL21 p 5 0.35 exosomes. The panel is a representative of a single region of the MFD to indicate entire cell population (Hoechst nuclear stain) and Ki67 positive cells (pink stain). |
|
Journal Publications
|
|
Conference Presentations / Publications
|
- Mercurio, A. (SMART/Univ. of Bari), Giulia Adriani* (SMART), Roger Kamm (MIT/SMART), et al, "A Mini-Review on Thalidomide: Chemistry, Mechanisms of Action, Therapeutic Potential and Anti-Angiogenic Properties in Multiple Myeloma." Current medicinal chemistry (2017),
- Andrea Pavesi (SMART/A*STAR-IMCB), Anthony T Tan (Duke-NUS), S.Koh (A*STAR-SICS), M.Colombo, E.Antonecchia, Carlo M, Giulia Adriani (SMART), M.T.Raimondi, R.D.Kamm (SMART/MIT), Antonio Bertoletti (Duke-NUS)/A*STAR-SICS), A 3D microfluidic model for preclinical evaluation of TCR-engineered T Cells against solid tumors, JCI Insight, June (2017)
- Ho YT (NUS/SMART), Adriani G (SMART), Beyer S (SMART), Nhan PT (NUS), Kamm RD (MIT/SMART), Kah JCY (NUS), "A Facile Method to Probe the Vascular Permeability of Nanoparticles in Nanomedicine Applications", Scientific Reports, 7, 707 (2017)
- Ugolini*, Andrea Pavesi (SMART/A*STAR-IMCB), Rasponi*, Fiore*, Roger Kamm (MIT/SMART), Soncini* (*Politecnico di Milano), "Human cardiac fibroblasts adaptive responses to controlled combined mechanical strain and oxygen changes in vitro", Elife (published online), 18th March 2017
- Arora, Natasha (MIT), Alsous, Jasmin Imran (MIT), Guggenheim, Jacob W.* (MIT), Mak, Michael (MIT), Munera, Jorge (MIT), M. Wells James (MIT), Kamm R D (MIT/SMART), Asada, H. Harry (MIT/SMART), Shvartsman, Stanislav Y (MIT), and Griffith, Linda G (MIT) “A process engineering approach to increase organoid yield”, Development, 144(6), 1128-1136 (2017)
- Giulia Adriani (SMART), Dongliang Ma (Duke-NUS, Andrea Pavesi (SMART), Roger D. Kamm (MIT/SMART) and Eyleen L. K. Goh (Duke-NUS/YLL Medicine, NUS), "A 3D neurovascular microfluidic model consisting of neurons, astrocytes and cerebral endothelial cells as a blood–brain barrier", Lab Chip, 17, 448 (2017)
- Giulia Adriani (SMART), Andrea Pavesi (SMART), Anthony T.Tan (Duke-NUS), Antonio Bertoletti (Duke-NUS), J.P.Thiery (NUS-YLSchool of Medicine), Roger Kamm (MIT/SMART), "Microfluidic models for adoptive cell-mediated cancer immunotherapies", Drug Discovery Today, doi:10.1016/j.drudis.2016.05.006 (2016)
- Andrea Pavesi (SMART), Giulia Adriani (SMART), Andy Tay (NUS), Majid Ebrahimi Warkiani (UNSW, Australia), Wei Hseun Yeap (A*STAR-SIgN), Siew Cheng Wong (A*STAR-SIgN), Roger D. Kamm (MIT/SMART), "Engineering a 3D microfluidic culture platform for tumor-treating field application", Scientific Reports | 6:26584 | DOI: 10.1038/srep26584 (2016)
- Giulia Adriani (SMART), Jing Bai (SMART), Siew-Cheng Wong (A*STAR-SIgN), Roger D. Kamm (MIT/SMART), Jean Paul Thiery (NUS/A*STAR-IMCB), "M2a macrophages induce contact-dependent dispersion of carcinoma cell aggregates", Macrophage 2016; 3: e1222. doi: 10.14800
- Giovanni Stefano Ugolini, Marco Rasponi, Andrea Pavesi (SMART), Rosaria Santoro, Roger Kamm (MIT/SMART), Gianfranco Beniamino Fiore, Maurizio Pesce, Monica Soncini, "On-Chip Assessment of Human Primary Cardiac Fibroblasts Proliferative Responses to Uniaxial Cyclic Mechanical Strain", Biotechnology and BioEnginering, 113 (4), 859-869, 2016
- J Bai (SMART/MIT), TY Tu (SMART), C Kim (MIT), JP Thiery (NUS & A*STAR/IMCB)), RD Kamm (MIT/SMART). "Identification of drugs as single agents or in combination to prevent carcinoma dissemination in a microfluidic 3D environment", Oncotarget, 6(34):36603-14 (2015)
- Giovanni Stefano Ugolini, Marco Rasponi, Andrea Pavesi, Rosaria Santoro, Roger Kamm, Gianfranco Beniamino Fiore, Maurizio Pesce, Monica Soncini. On-chip assessment of human primary cardiac fibroblasts proliferative responses to uniaxial cyclic mechanical strain. Biotechnology and Bioengineering, Published online, Oct 2015
- Kim M-C (MIT), Whisler J (MIT), Silberberg YR (SMART), Kamm RD (MIT/SMART), Asada HH (MIT/SMART), "Cell Invasion Dynamics into a Three Dimensional Extracellular Matrix Fibre Network", PLoS Comput Biol 11(10): e1004535. doi:10.1371/journal.pcbi.1004535 (2015)
- Jing Bai (SMART), Giulia Adriani (SMART), Truong-Minh Dang (SIgN-A*STAR), Ting-Yuan Tu (SMART), Hwei-Xian Leong Penny (SIgN-A*STAR), Siew-Cheng Wong (SIgN-A*STAR), R.D. Kamm (MIT/SMART) and Jean-Paul Thiery (NUS/IMCB-A*STAR/SMART), "Contact-dependent carcinoma aggregate dispersion by M2a macrophages via ICAM-1 and beta2 integrin interactions", Oncotarget, Advance Publications 2015
- Andrea Pavesi (SMART), Giulia Adriani (SMART), Marco Rasponi (Politecnico di Milano, Italy), Ioannis K. Zervantonakis (MIT), Gianfranco B. Fiore ( (Politecnico di Milano, Italy) and Roger D. Kamm (MIT / SMART), "Controlled electromechanical cell stimulation on-a-chip" Scientific Reports, 5, 11800 (2015)
- Young K. Park (SMART), Ting-Yuan Tu (NUS/SMART), Sei Hien Lim (NUS/SMART), Ivan J. M. Clement (NUS/SMART), Se Y. Yang (MIT), Roger D. Kamm (MIT/SMART), "In Vitro Microvessel Growth and Remodeling within a Three-Dimensional Microfluidic Environment", Cellular and Molecular Bioengineering, 7 (1), 15 (2014)
- Soheila Sharghi-Namini (SMART), Evan Tan (NUS/SMART), Lee-Ling Sharon Ong (SMART), Ruowen Ge (NUS)and H. Harry Asada (MIT/SMART), "Dll4-containing exosomes induce capillary sprout retraction in a 3D microenvironment", SCIENTIFIC REPORTS | 4 : 4031 | DOI: 10.1038/srep04031 (2014)
- Sahan Herath (NUS/SMART), Du Yue (NUS/SMART), Dong-an Wang (NTU), Kin Liao (NTU), Qingguo Wang (NUS), Harry Asada (MIT/SMART) and Peter Chen (NUS), "Characterization of uniaxial stiffness of extracellular matrix embedded with magnetic beads via bio-conjugation and under the influence of an external magnetic field", Journal of the Mechanical Behavior of Biomedical Materials, 30, 253 (2014)
- Sahan Herath (NUS/SMART), Du Yue (NUS/SMART), Shi Hui (SMART), Min-Cheol Kim (SMART), Dong-an Wang (NTU), Qingguo Wang (NUS), Krystyn Van Vliet (MIT/SMART), Harry Asada (MIT/SMART) and Peter Chen (NUS), "Quantification of Magnetically Induced Changes in ECM Local Apparent Stiffness", BioPhysical Journal, 106, 332-341 (2014)
- C. J. Ochs (SMART), J. Kasuya (MIT) , A. Pavesi (SMART) and R.Kamm (MIT/SMART), “Oxygen Levels in Thermoplastic Microfluidic Devices during Cell Culture”, Lab on A Chip, 2013, DOI: 10.1039/C3LC51160J
- Sei Hien Lim (NUS/SMART), Choong Kim (SMART), Amir R. Aref (SMART), Roger D. Kamm (MIT/SMART), Michael Raghunath (NUS), "Complementary effects of ciclopirox olamine, a prolyl hydroxylase inhibitor and sphingosine 1-phosphate on fibroblasts and endothelial cells in driving capillary sprouting", Integrative Biology (In Press), published online 30 Oct 2013
- Lee-Ling S. Ong (SMART), Justin Dauwels (NTU), Marcelo H. Ang Jr. (NUS) and H. Harry Asada (MIT/ SMART), “A Bayesian Filtering Approach to Incorporate 2D/3D Time-lapse Confocal Images for Tracking Angiogenic Sprouting Cells Interacting with the Gel Matrix”, Medical Image Analysis, 2013, doi: 10.1016/j.media.2013.10.008
- X.M. Ang (NUS), M.H.C. Lee (NUS), A. Blocki (NUS), C. Chen (NUS), L.L.S Ong (SMART), H.H. Asada (MIT/SMART), A. Sheppard (University of Auckland), M. Raghunath (NUS), “Macromolecular crowding amplifies adipogenesis of human bone marrow-derived MSCs by enhancing the pro-adipogenic microenvironment”, Tissue Engineering A, 2013, doi: 10.1089/ten.TEA.2013.0337
- Ting-Yuan Tu (NUS/SMART), Zhe Wang (NUS/SMART), Jing Bai (SMART), Wei Sun (SMART), Weng Kung Peng (SMART), Ruby Yun-Ju Huang (CSIS/NUS), Jean-Paul Thiery (NUS/A*STAR-IMCB), Roger D. Kamm (MIT/SMART), "Rapid Prototyping of Concave Microwells for the Formation of 3D Multicellular Cancer Aggregates for Drug Screening", Advanced Healthcare Materials, Published online, Aug 2013 (DOI: 10.1002/adhm.20130015, © 2013 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim
- Min-Cheol Kim (SMART), Raymond H. W. Lam (MIT), Todd Thorsen (MIT), and H. Harry Asada (MIT/SMART), "Mathematical Analysis of Oxygen Transfer through Polydimethylsiloxane Membrane between Double Layers of Cell Culture Channel and Gas Chamber in Microfluidic Oxygenator", Microfluidics and Nanofluidics, (Published online), DOI 10.1007/s10404-013-1142-8, Feb 1, 2013
- Min-Cheol Kim (SMART), Devin Neal (MIT), Roger D. Kamm (MIT/SMART), and H. Harry Asada (MIT/SMART), "Dynamic Modeling of Cell Migration and Spreading Behaviors on Fibronectin Coated Planar Substrates and Micropatterned Geometries", PLOS Computational Biology, 9(2), e1002926, 2013
- Amir R. Aref (SMART), Ruby Yun-Ju Huang (CSIS/NUS), Wei-miao Yu (A*STAR-IMCB), Kian-Ngiap Chua (CSIS-NUS)),
Wei Sun (SMART), Ting-Yuan Tu (NUS/SMART), Jing Ba (SMART) , Wen-Jing Sim (CSIS-NUS)), Ioannis K Zervantonakis (MIT), Jean Paul Thiery (A*STAR-IMCB) and Roger D. Kamm (SMART/MIT), "Screening therapeutic EMT blocking agents in a three dimensional microenvironment" Integrative Biology, published on the web 21 Nov 2012, DOI: 10.1039/C2IB20209C
- Kenichi Funamoto (Tohoku Univ), Ioannis K.Zervantonakis (MIT), Liu, Y. (NUS), Ochs, C. J. (SMART), Kim, C. (SMART), Kamm, R (MIT/SMART), "A Novel Microfluidic Platform for High-Resolution Imaging of a Three-Dimensional Cell Culture under a Controlled Hypoxic Environment", Lab Chip, 22, 4855-4863 (2012).
- Kim, M. (SMART), Kim, C. (SMART), Wood, L. (MIT), Neal, D. (MIT), Kamm, R. (MIT / SMART) and Asada, H. (MIT / SMART), "Integrating Focal Adhesion Dynamics, Cytoskeleton Remodeling, and Actin Motor Activity for Predicting Cell Migration on 3D Curved Surfaces of Extracellular Matrix", Integrative biology, 4(11), 1386-1397 (2012)
- Vickerman V (MIT) and Kamm RD (MIT/SMART), "Mechanism of a flow-gated angiogenesis switch: early signaling events at cell-matrix and cell-cell junctions", Integr. Biol (Camb). 2012 Aug 23; 4(8):863-74. Epub 2012
- Wood,
L.(MIT),
Ge,
R.
(NUS),
Kamm,
R.
(MIT/SMART),
and
Asada,
H.H.
(MIT/SMART), “Nascent Vessel
Elongation
rate
is
Inversely Related
to
Diameter
in
In Vitro
Angiogenesis,
Integrative
Biology.” Integrative
Biology, 4, 1081 (2012)
- Choong Kim (SMART), Seok Chung (Korea Univ), Liu Yuchun (NUS), Min-Cheol Kim (SMART), Jerry Chan (Duke-NUS), H. Harry Asada (MIT/SMART), and Roger D. Kamm (MIT/SMART), "In vitro angiogenesis assay for the study of cell-encapsulation therapy", Lab on A Chip, 12(16), 2942 (2012)
- Ge, R. (NUS), Tan, E. (NUS/SMART), Sharghi-Namini, Soheila SMART), Asada, H. (MIT/SMART), “Exosomes in Cancer Microenvironment and Beyond: Have We Overlooked These Extracellular Messengers?”, Cancer Microenvironment, Vol. 5, DOI 10.1007/s12307-012-0110-2, May 2012.
- Farahat, W. (MIT), Wood, L. (MIT), Zervantonakis, I. (MIT), Schor, A. (MIT), Ong, S. (SMART), Neal, D. (MIT), Kamm, R. (MIT/SMART), and Asada, H. (MIT/SMART), “Ensemble Analysis of Angiogenic Growth in Three-Dimensional Microfluidic Cell Cultures”, PLoS ONE 7(5): e37333. doi:10.1371/journal.pone.0037333, May 2012.
- H.K. Taylor (SMART), "Simulation and mitigation of process and pattern dependencies in nanoimprint lithography", Journal of Photopolymer Science and Technology, vol. 24, no. 1, pp. 47-55, 2011.
- Polacheck WJ (MIT), Charest JL (MIT), Kamm RD (MIT/SMART), "Interstitial flow influences direction of tumor cell migration through competing mechanisms", PNAS, 2011 Jun 20. PubMed PMID: 21690404
- Wood, L., Kamm, R., Asada, H., "Stochastic Modeling and Identification of Emergent Behaviours of an Endothelial Cell Population in Angiogenic Pattern Formation", International Journal of Robotics Research (Published online, 11th March 2011).
- C.R.Wan, Seok Chung and Roger D. KAMM, "Differentiation of Embryonic Stem Cells into Cardiomyocytes in a Compliant Microfluidic System", Annals of Biomedical Engineering, DOI: 10.1007/s10439-011-0275-8 (Published online in Feb. 2011)
- C.R.Wan, E.M.Frohlich, J.L.Charest and R.D.Kamm, "Effect of Surface Patterning and Presence of Collagen I on the Phenotypic
Changes of Embryonic Stem Cell Derived Cardiomyocytes", Cellular and Molecular Bioengineering, 4, 56 (2011)
- Zervantonakis IK, Chung S, Sudo R, Zhang M, Charest JL, Kamm RD. "Concentration gradients in microfluidic 3D matrix cell culture systems" Intern J Micro-Nano Scale Transport:, 1(1), 27 (2010)
- Zhang, W., Tucker-Kellogg, L., Narmada, B.C., Venkatraman, L., Chang, S., Yin, L., and Yu, H. "Cell-Delivery Therapeutics for Liver Regeneration", Advanced Drug Delivery Reviews, published online, March 1, 2010
- Wood, L., Das, A., Kamm, R. and Asada, H., "A Stochastic Broadcast Feedback Approach to Regulating Cell Population Morphology for Microfluidic Angiogenesis Platforms", IEEE Transactions on Biomedical Engineering, 56(9), Part 2, 2299 (2009)
- Chung S, Sudo R, Zervantonakis I, Rimchala T, Kamm RD. "Surface-treatment-induced three-dimensional capillary morphogenesis in a microfluidic platform" (Cover Article) Advanced Materials, 21(47), 4863 (2009)
- Lee H, Pelz B, Ferrer JM, Kim T, Lang MJ, and Kamm RD, "Cytoskeletal Deformation at High Strains and the Role of Cross-link Unfolding or Unbinding", Cellular and Molecular Bioengineering, 2(1), 28 (2009)
- Chung S, Yun H, and Kamm RD, "Nanointerstice-driven microflow", Small, 5(5), 609 (2009)
- Sudo R, Chung S, Zervantonakis IK, Vickerman V, Toshimitsu Y, Griffith LG, Kamm RD, "Transport-mediated angiogenesis in 3D epithelial co-culture", FASEB J, 23, 1 (2009)
- Zhang, C., Zhao, Z., Abdul Rahim, N.A., van Noort, D., and Yu, H "Towards a Human-on-Chip: Culturing Multiple Cell Types on a Chip with Compartmentalized Microenvironments", Lab on a chip, 9(22), 3185 (2009)
|
|
- Giulia Adriani (SMART), Hweixian Leong Penny (A*STAR-SIgN), Je Lin Sieow (A*STAR-SIgN), Siew Cheng Wong (A*STAR-SIgN), Roger D. Kamm (MIT/SMART), "Exploring the role of tumor-conditioned macrophage metabolism on extravasation of pancreatic ductal adenocarcinoma cells", AACR Annual Meeting, New Orleans, USA, May 2016
- G. Adriani (SMART), D. Ma (Duke-NUS), A. Pavesi (SMART), E.L. Goh (Duke-NUS), R.D. Kamm. “Modeling the blood-brain barrier in a 3D triple co-culture microfluidic system”, (Invited), 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Milan, Italy, Aug 2015.
- Pavesi (SMART), T.A. Tanoto (Duke-NUS), M. Chen (MIT), G. Adriani (SMART), B. Antonio (Duke-NUS), R.D. Kamm (MIT), “Using Microfluidics to Investigate Tumor Cell Extravasation and T-Cell Immunotherapies”, (Invited) 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Milan, Italy, Aug 2015
- Mengmeng Wang (NTU), Lee-Ling S. Ong (SMART), Justin Dauwels (NTU) and H. Harry Asada (MIT), "Automated Tracking of Cells from Phase Contrast Images by Multiple Hypothesis Kalman Filters." IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), April - 2015, pp. 942-946.
- S. Vaikundam (NTU), L.L.S Ong (SMART), J. Dauwels (NTU), H. H. Asada (MIT), "Automated tracking of collagen fibers from confocal reflectance images", at the International Conference on Optical and Photonic Engineering (icOPEN 2015), Paper 9524-160, 14-16 April 2015.
- M. Mathew (NTU), LLS Ong (SMART), J. Dauwels (NTU), H. H. Asada (MIT), "Automated segmentation and classification of unlabelled malaria parasites in red blood cells from phase contrast images", at the International Conference on Optical and Photonic Engineering (icOPEN 2015), Paper 9524-158, 14-16 April 2015.
- Mengmeng Wang (SMA3/NTU), Lee-Ling S. Ong (SMART), Justin Dauwels (NTU) and H. Harry Asada (MIT), "Automatic Detection of Endothelial Cells in 3D Angiogenic Sprouts from Experimental Phase Contrast Images." In SPIE Medical Imaging, pp. 94132I-94132I, Feb 2015, Florida, USA
- A. Pavesi (SMART), G. Adriani (SMART), M. Rasponi (Politecnico di Milano, ITALY), G.B. Fiore (Politecnico di Milano, ITALY), I. Zervantonakis (MIT)and R.D. Kamm (MIT/SMART), "Lab-on-a-chip based microfluidic environment for cell electromechanical stimulation", Proceedings of the 6th International Symposium on Microchemistry and Microsystems (ISMM), pp 37-38, 30 July – 1 August 2014,
Singapore
- Y. R. Silberberg (SMART), M. Kim (SMART), H. H. Asada (MIT/SMART), "3D Traction Force Microscopy Combined with Computer Simulation Models for the Analysis of Filopodia Dynamics", 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- V. Rajagopal (SMART), A. Pavesi (SMART), W. Polacheck (MIT), R. D.
Kamm (MIT/SMART), "Quantification of the Spatial Distribution of Cytoskeletal Proteins and Adhesions of MDA-MB-231 Breast Cancer Cells Embedded inside 3D Collagen Matrices", 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- S. Beyer (SMART), Q. Liu (SMART), G. Adriani (SMART), A. M. Blocki (A*STAR-SBIC), J.
Yang (NUS), J. Chan (Duke-NUS/KKH), R. D. Kamm (MIT/SMART), "The role of the glycocalyx and heparan sulfates within the basement membrane of human microvasculature in transendothelial migration", 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- M. Mayalu (MIT), M. Kim (SMART), V. Chan (MIT), D. Neal (MIT), H. Kim (MIT),
H. Asada (MIT/SMART), "A New Approach to Predicting Emergent Behaviors of Cell-ECM Interactions Using Stochastic Rules Generated from Mechanistic Computational Models, 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- A. Pavesi (SMART), T. A. Tanoto (Duke-NUS), A. Bertoletti (Duke-NUS), R. D.
Kamm (MIT/SMART), "Interactions Between Hepatocellular Carcinoma Cells and T-cell Receptor Engineered T Cells in a
3D Microfluidic Platform", 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- M. Kim (SMART), R. Kamm (MIT/SMART), H. Asada (MIT/SMART), "Filopodia dynamics, ECM interaction, and 3D cell migration modeling", 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- A. Pavesi (SMART), C. J. Ochs (SMART), J. Kasuya (MIT), R. D. Kamm (MIT/SMART), "Oxygen Level in Thermoplastic Microfluidic Devices", 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- S. Huang (NUS/SMART), J. Chan (Duke-NUS/KKH), H. Yu (NUS), R. D. Kamm (MIT/SMART), "Neural Stem Cells Homing to Glioma Vasculature", 7th World Congress of Biomechanics, July 6-11, 2014, Boston, MA
- Min-Cheol Kim (SMART), Chin Wen Tan (SMA3/NUS), Jerry Chan (Duke-NUS/KKH), Linda G. Griffith (MIT/SMART), Roger D. Kamm (MIT/SMART), H. Harry Asada (MIT/SMART), “Integrated Cell Migration Model incorporating Spatiotemporal Kinetics of Focal Adhesion Assembly and Disassembly”, BMES 2013 Annual Meeting, Sep 25-28, 2013, Seattle, Washington.
- Min-Cheol Kim (SMART), Peter C. Y. Chen (NUS), Roger D. Kamm (MIT/SMART), H Harry Asada (MIT/SMART), “Incorporating Filopodia dynamics, Focal adhesion dynamics, Cytoskeleton remodeling, and Degradation of Extracellular matrix for Predicting Tip Cell Migration in Angiogenesis”, BMES 2013 Annual Meeting, Sep 25-28, 2013, Seattle, Washington.
- Baijing (SMART), T.Y. Tu (NUS/SMART), J.P. Thiery (NUS), and R.D. Kamm (MIT/SMART), "Microfluidic 3D cancer-type specific platform for EMT blocking agents screening", BMES-2013, USA.
- T-Y. Tu (SMART/MBI/NUS), J. Bai (SMART), R. Huang (NCC), J-P. Thiery (A*STAR IMCB/NUS), and R. D. Kamm (MIT/SMART), "A Microfluidic Co-culture System for Epithelial-Mesenchymal Transition Drug Screening", Biomedical Engineering Society (BMES) Meeting, Atlanta, GA, Oct-2012
- M-C. Kim (SMART), C. Kim (SMART), L. Wood (MIT), D. Neal (MIT), R. Kamm (SMART/MIT) and H. Asada (SMART/MIT), "2-D Cell Migration on the 3-D Curved Surfaces: Experiment and Simulation", Biomedical Engineering Society (BMES) Meeting, Atlanta, GA, Oct-2012
- M-C. Kim (SMART), R. Kamm (SMART/MIT), and H. Asada (SMART/MIT), "Introduction of Integrated Cell migration Model: Comparisons with Cell Migration Experiments", BioMedical Engineering Society, BMES-2012, Atlanta, GA, USA
- Sei Hien Lim (SMA3/NUS), Amir R. Aref (SMART), Choong Kim (SMART), Michael Raghunath (NUS), Roger D. Kamm (MIT/SMART), "Induction of Angiogenesis in microfluidic devices using prolyl hydroxylase inhibitors and sphingosine-1 phosphase", MicroTAS-2012, Oct., 2012
- Yie Hou Lee (SMART), “Targeted sphingolipidomics reveal dysregulated glucosylceramide levels in endometriosis”Invited talk, 16th World Congress on Controversies in Obstetrics, Gynecology & Infertility (COGI). Singapore. July 2012
- Christopher J. Ochs (SMART), Roger Kamm (MIT/SMART), D.Trau (NUS), “Oxygen Sensors for Microfluidic 3D Cell Culture", Europtrode XI, Barcelona, April 2012
- C.W. Tan (NUS/SMART), S.F. Loh (KKH) , H.H. Tan (KKH), M. Choolani (NUS), L. Griffith (MIT/SMART), J. Chan (NUS),"Endometrial cell motility in endometriosis under hypoxic microenvironment", 2012 Society of Gynecologic Investigation 59th Annual Scientific Meeting, San Diego, USA, March 22-25, 2012.
- Christopher J. Ochs (SMART), "Sensing of Biochemical Cues in 3D Cell Cultures”, 2nd Molecular Materials Meeting, Singapore, January-2012
- Christopher J. Ochs (SMART), "Sensors for Microfluidic 3D Cell Cultures”, MBI Workshop on Forces and Cells, Singapore, November-2011
- Y. K. Park (SMART), C. S. Khoon (NUS), T-Y. Tu (NUS), I. J. Clement (NUS), and R. D. Kamm (MIT/SMART), "Three Dimensional Microfluidic System for Screening Anti-Metastatic Cancer Drugs", Annual Meeting of Biomedical Engineering Society (BMES) @ Connecticut, USA, Oct 2011
- T.Y. Tu (NUS), W. Sun (SMART), W.K. Peng (SMART), Y.J. Huang (NUS), P.T. Matsudaira (NUS), J.P. Thiery (A*STAR IMCB), and R.D. Kamm (MIT/SMART), "Rapid microell prototyping, generation of 3D cancer spheroids and EMT drug screening", MicroTAS 2011, Seattle, USA, October 2-6, 2011, pp. 82-84.
- Choong Kim (SMART), Seok Chung (Korea Univ.), Min-Cheol Kim (SMART), and Roger D. Kamm (MIT/SMART), “A new in vivo-mimic angiogenesis microfluidic platform for the study of cell encapsulation based cell therapy”, MicroTAS 2011, Seattle, USA, October 2-6, 2011, pp. 783-785.
- Choong Kim (SMART, KIST), Jae Hoon Bang (KIST), Young Eun Kim (KIST), and Ji Yoon Kang (KIST), "Microfluidic manifold system to reduce the number of syringe pumps in multi-phase systems for generating alginate beads", MicroTAS 2011, Seattle, USA, October 2-6, 2011, pp. 266-268.
- Wood, L. (MIT) and Asada, H. (MIT/SMART), “An Experimentally Tuned Dynamic Model Predicting Cell Migration for Guidance of Sprouting Endothelial Cells”, Proceedings of Dynamic Systems and Control Conference, Arlington, VA, Oct/Nov, 2011.
- Yuchun Liu (NUS), Jerry Chan (Duke-NUS/KKH), Roger Kamm (MIT/SMART), Mark SK Chong (NUS), Zhang Zhiyong (NUS), Mahesh A Choolani (NUS), Swee-Hin Teoh (NUS), "Generating Vascularised Tissue-Engineered Bone Grafts: Endothelial Progenitor Cells in Vasculogenic and Osteogenic Priming of Human Fetal Mesenchymal Stem Cells", The International Bone-Tissue-Engineering Congress, Hannover, Oct-2011
- C.W. Tan (NUS/SMART), S.F. Loh (KKH), H.H. Tan (KKH), M. Choolani (NUS), L. Griffith (MIT/SMART), J. Chan (Duke-NUS/KKH), "Migratory behaviour of cells derived from eutopic endometrium in endometriosis under hypoxic microenvironment”, World Congress of Endometriosis 2011 (Sept-2011), Montpellier, France, & 8th Singapore International Congress of Obstetrics and Gynaecology, Singapore, August 25-27 2011
- Ramanathan, K. (NUS), Ong, L.L.S (SMART), Asada, H. H. (MIT/SMART), Ang, M. H. (NUS), “Automated tracking of biological cells in an “in-vitro” environment using active contours and distance measures,” Proceedings of the IEEE 5th International Conference on Cybernetics and Intelligent Systems (CIS), 2011, Pages 241—246, Qingdao, China, 17—19 September, 2011.
- Lee-Ling S. Ong (SMART), Levi B. Wood (MIT), Marcelo H. Ang Jr. (NUS) and H. Harry Asada (MIT/SMART), “Stochastic Tracking of Migrating Live Cells Interacting with 3D Gel Environment Using Augmented-Space Particle Filters”, IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), Sept-2011, San Francisco, CA, USA.
- C.W. Tan (NUS/SMART), S.F. Loh (KKH) , H.H. Tan (KKH), M. Choolani (NUS), L. Griffith (MIT/SMART), J. Chan (NUS),"Clonogenicity of Stromal/ Epithelial Cell- like Stem Cells Derived from Eutopic Endometrium in Endometriosis", 27th Annual Meeting of the European Society of Human Reproduction & Embryology, Stockholm, Sweden, July 3-7 2011.
- Peter Chen (NUS), Sahan Herath (NUS), Dongan Wang (NTU), Su Kai (NTU), Liao Kin (NTU) and Harry Asada (MIT/SMART),"Active Manipulation of ECM Stiffness", ASME Summer Bioengineering Conference, June-2011, PA, USA.
- S.H. Lim, A.R. Aref, M. Raghunath, R.D. Kamm, “Induction of Angiogenesis in Microfluidic Devices using Prolyl Hydroxylase Inhibitors,” 2nd Conference on Advances in Microfluidics and Nanofluidics and Asian Pacific International Symposium on Lab on chip (AMN-APLOC), Jan 2011, Singapore
- M. Kim, C. Klapperich and H. Asada, "Numerical Methods for Simulating Biconcave Cells in the Microfluidic Device", The 2nd conference on advanced in microfluidics and nanofluiducs 2011, Jan 2011, Singapore.
- A.R.Aref, R.Y.J. Huang (IMCB), W. Kang, S.W. Jing, J.P.Thiery (IMCB), R.D.Kamm, "Microfluidic Co-culture Platform for Studying EMT in 3D Scaffolds", 14th International Biotechnology Symposium and Exhibition, 14-18 September 2010, Rimini, Italy.
- Young K. Park, Sharon L. L. Ong, Devin Neal, Mayasari Lim, and Roger D. Kamm, " In vitro preclinical microfluidic screening for drug-like molecules as cardiomyocyte differentiation agents" 2nd International Conference on Cellular and Molecular Bioengineering (ICCMB2), August 2-4, 2010 in NTU Singapore
- Roger Kamm, "Integrated cellular systems: A bioengineering approach to the design of biological machines", 6th World Congress of Biomechanics, 1-6 August 2010, Singapore, Proceedings pp.174
- Amir Aref, Ruby Huang, Jean Paul Thiery and Roger Kamm, "Transitions between epithelial and mesenchymal states in Microfluidic platform: Aquisition of malignant and stem cell traits", 6th World Congress of Biomechanics, 1-6 August 2010, Singapore, Proceedings pp.553-554
- Hossein Nejadnik, Amir Aref, James Hui Hoi Po and Roger Kamm, "Simulating injured cartilage environment by microfluidic platform for mesenchymal stem cell homing evaluation", 6th World Congress of Biomechanics, 1-6 August 2010, Singapore, Proceedings pp.559
- A.R. Aref, R.Y.J. Huang, W. Kang, S.W. Jing, J.P. Thiery, R.D. Kamm, "Transitions Between Epithelial and Mesenchymal States in Microfluidic Platform: Acquisition of Malignant and Stem Cell Traits", TechConnect World Conference 2010, World Conference and Expo 2010, June 21-25, 2010 in Anaheim, California, U.S.A.
- Wood, L., Kamm, R., Asada, H., "Time Lapse Observation Based Modeling and Identification of Cell Behaviours in Angiogenic Sprout Development", Proceedings of the Dynamic Systems and Control Conference - 2010.
- Farahat, W. and Asada, H., "Control of Eukaryotic Cell Migration via Modulation of Extracellular Chemoattractant Gradients", Proceedings of the 3rd ASME Dynamical Systems and Control Conference, Cambridge, MA 2010.
- Farahat, W. and Asada, H., "Estimation of State Transition Probabilities in Asynchronous Population Markov Processes", Proceedings of the American Control Conference, Baltimore, MD, 2010.
- Lee-Ling S. Ong, Marcelo H. Ang Jr. and H. Harry Asada, "Tracking of Cell Population from Time Lapse and End Point Confocal Microscopy Images with Multiple Hypothesis Kalman Smoothing Filters", IEEE Computer Society Workshop on Mathematical Methods in Biomedical Image Analysis 2010, San Francisco, CA, USA, June 14 2010
- Waleed Farahat and Harry Asada, "Identification of phenotypic state transition probabilities in living cells", 2009 ASME Dynamic Systems and Control Conference
- Abdul Rahim, N.A., Iliescu, C., Kamm, R., Yu, H. “Microfluidic device captures correlation between human mesenchymal stem cell differentiation capacity and migration activity” World Stem Cell Summit, Baltimore, Maryland, USA, 21-23 September 2009
- Yu, H. (2009) “Engineering in vitro drug testing platforms” ICMAT Symposium A - Advanced Biomaterials and Regenerative Medicine (In Conjunction with 2nd Asian Biomaterials Congress), Singapore, 28 June – 3 July 2009
- Yue, Z., Mo, X., Nugraha, B., Tan, C.H., Yu, H. (2009) “Biomaterials to Facilitate Controls of Cell-Matrix and Cell-Cell Interactions in Soft Tissue Construction” 2nd Asian Biomaterials Congress (ABMC), Singapore, 26 - 27 June 2009
- Wood, L., Asada, H., "Stable Control of Distributed Hysteretic Systems Using Cellular Broadcast Stochastic Feedback", Proceedings of American Control Conference, Jun. 2009
|
|
 |