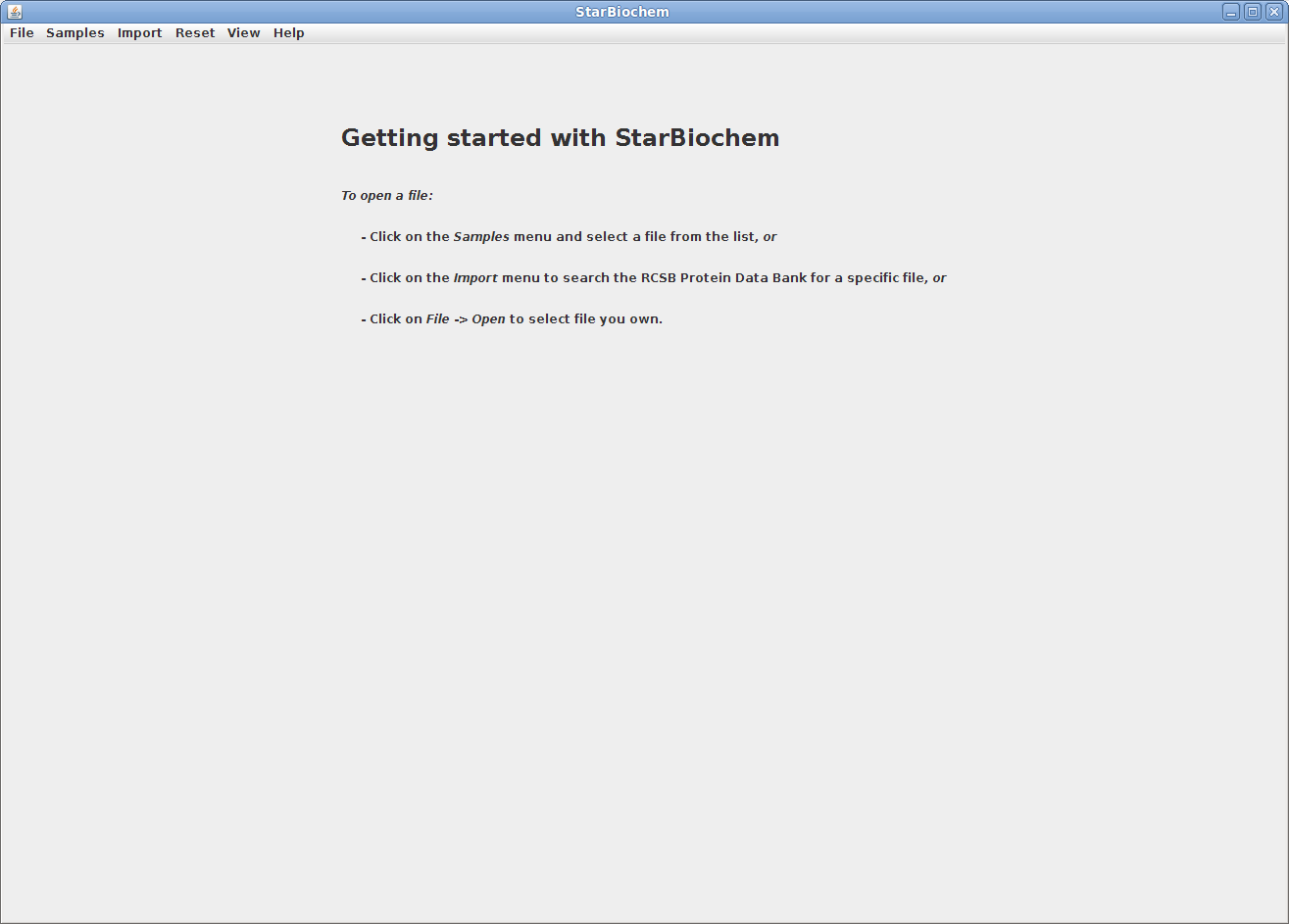
StarBiochem is a 3D protein viewer and runs on standard Linux, Windows and Mac computers.
To get started with StarBiochem:
Navigate to http://web.mit.edu/star/biochem.
Click on the Start button to launch the StarBiochem application.
This is the view you will see
StarBiochem opens.
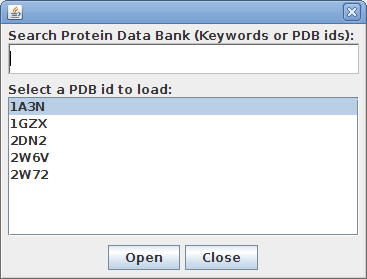
StarBiochem enables the visualization of molecules encoded within the Protein Data Bank (PDB) files. PDB files are named by a four-character ID such as “1GXX”. You may load any file found in the PDB, select one from the Samples menu or open files stored locally on your computer.
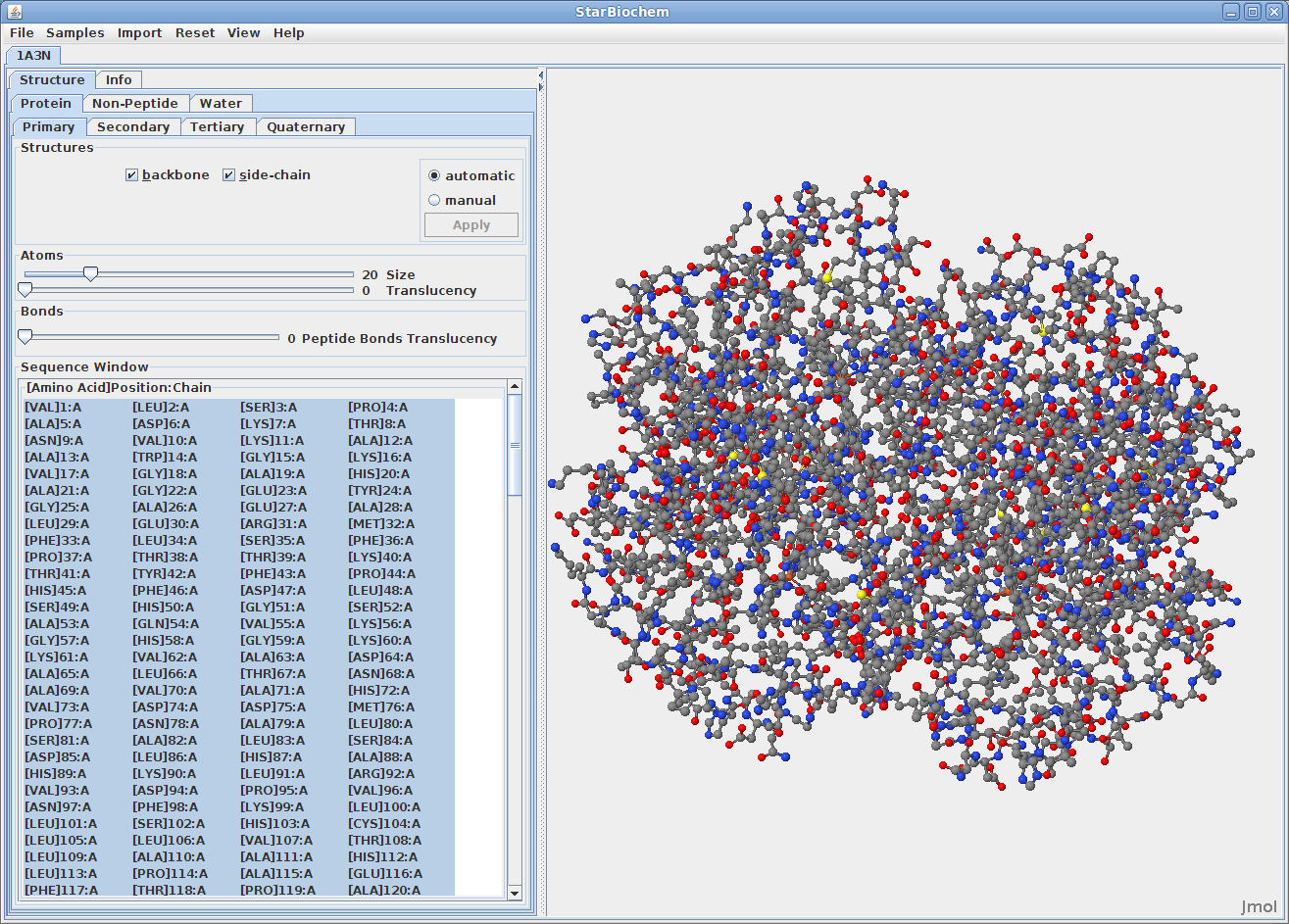
Upon opening a PDB file in StarBiochem, you will see a 3D model of the protein in the viewer.
When multiple PBD files are open, they can be closed
individually or all at once by clicking on
File -> Close, or File
-> Close
All respectively.
Note:
Each tab contains
Automatic and
Manual options
which allow you to choose when the changes made are
applied to the image.

In the top menu selectHelp->Structure viewing instructions. This will open a small window with instructions for manipulating the image of the structure.
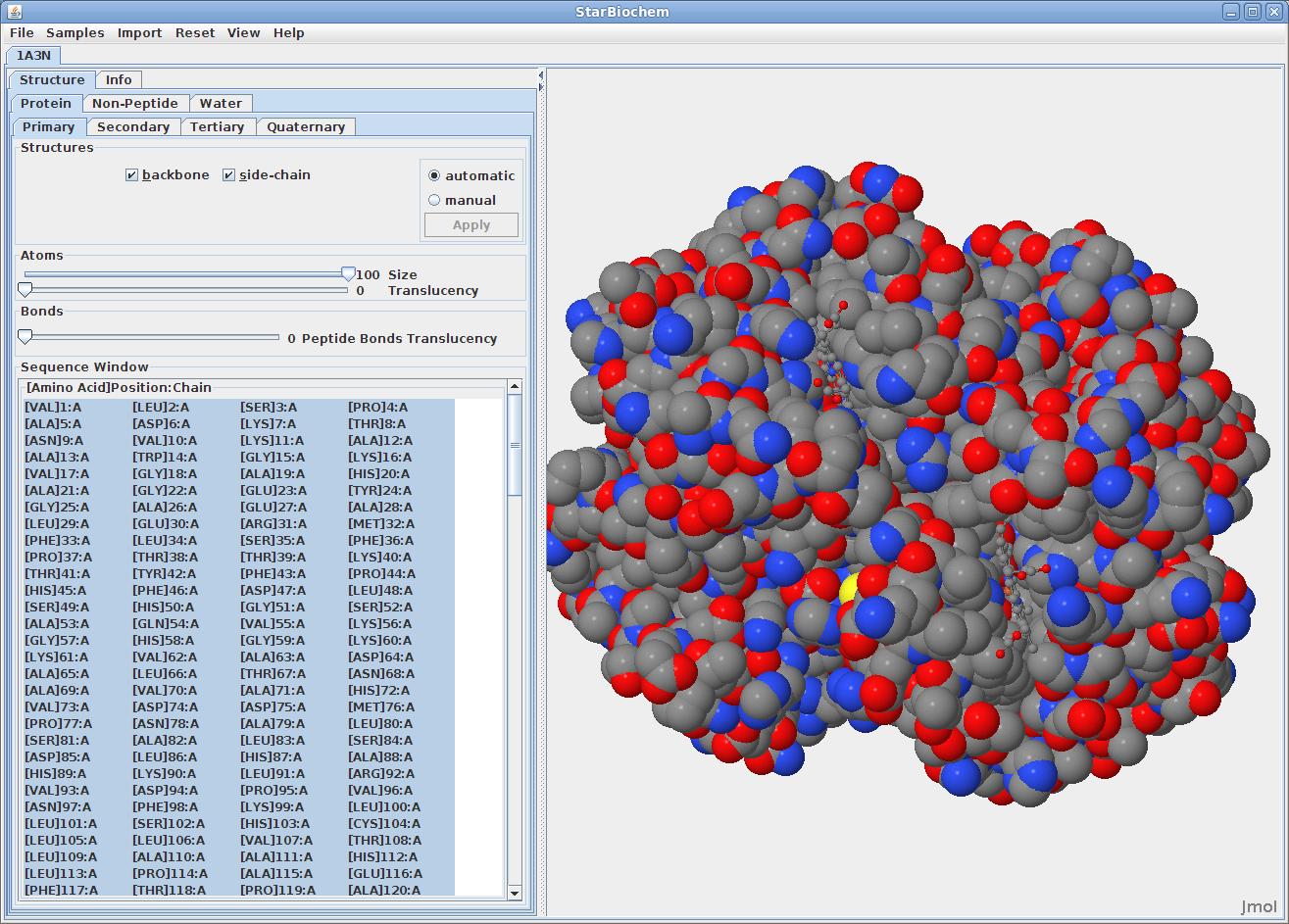
There are two different 3D models used to visually represent the structure of molecules:
You can switch from the ball-and-stick model to the space-filled model in StarBiochem by increasing the size render of the atoms in the structure.
The default coloring scheme for atoms is as
follows:
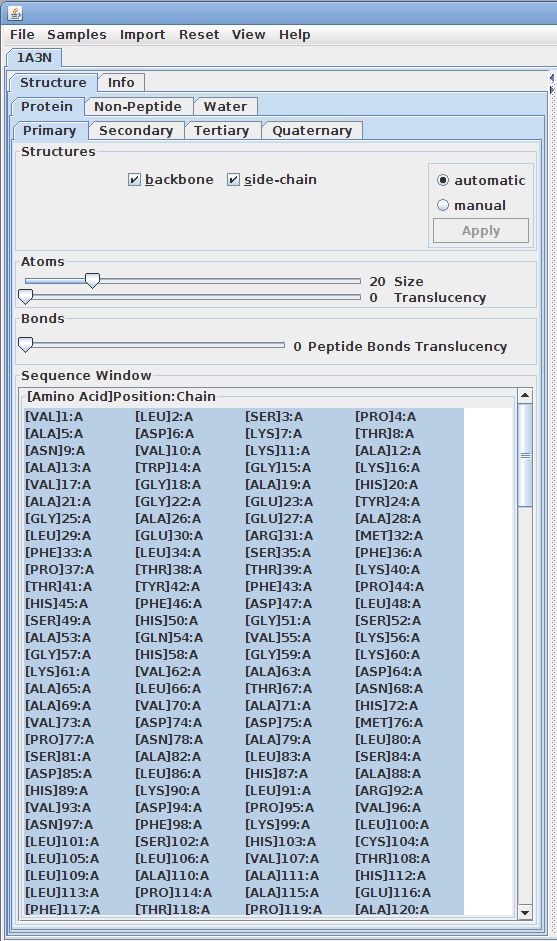
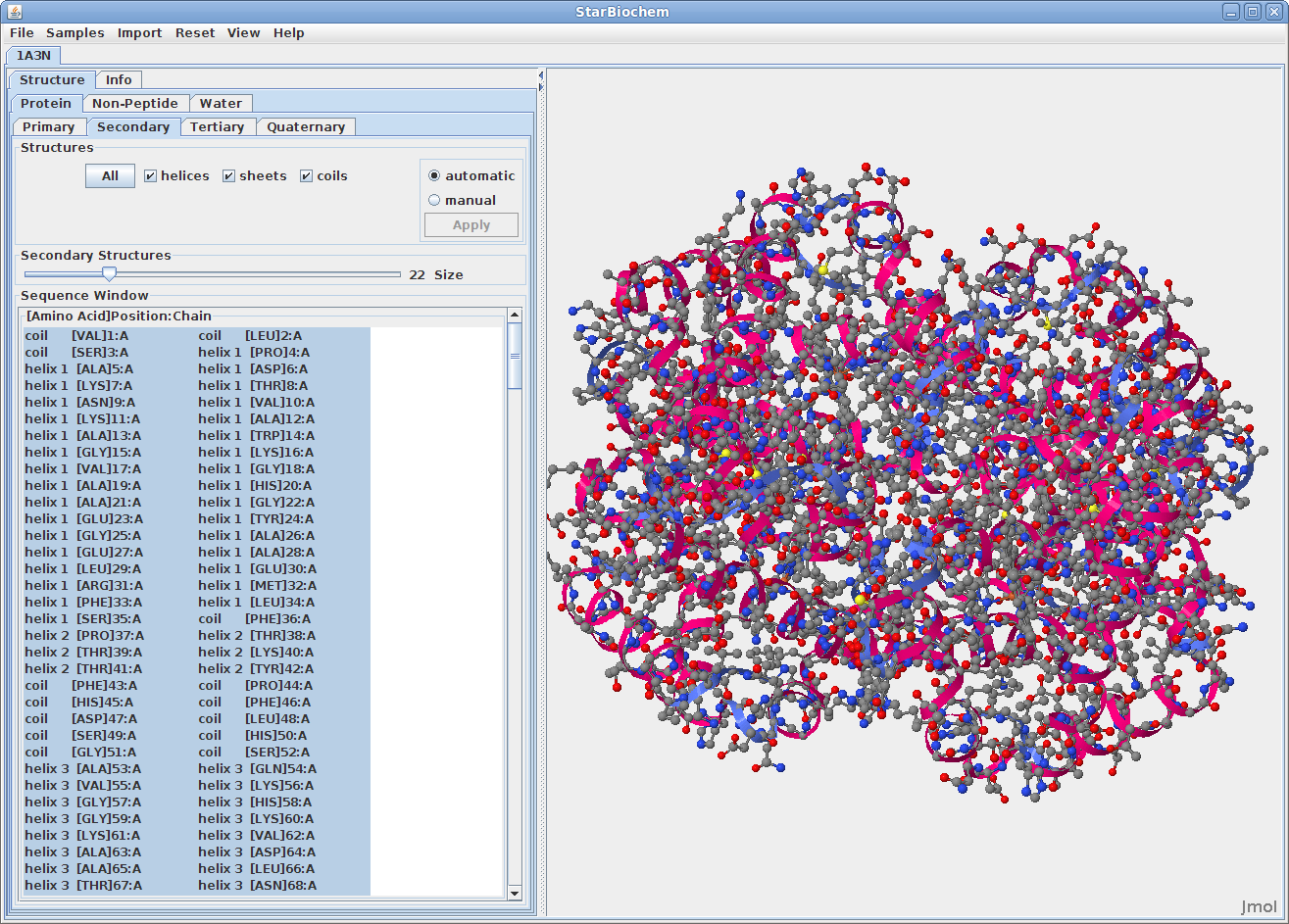
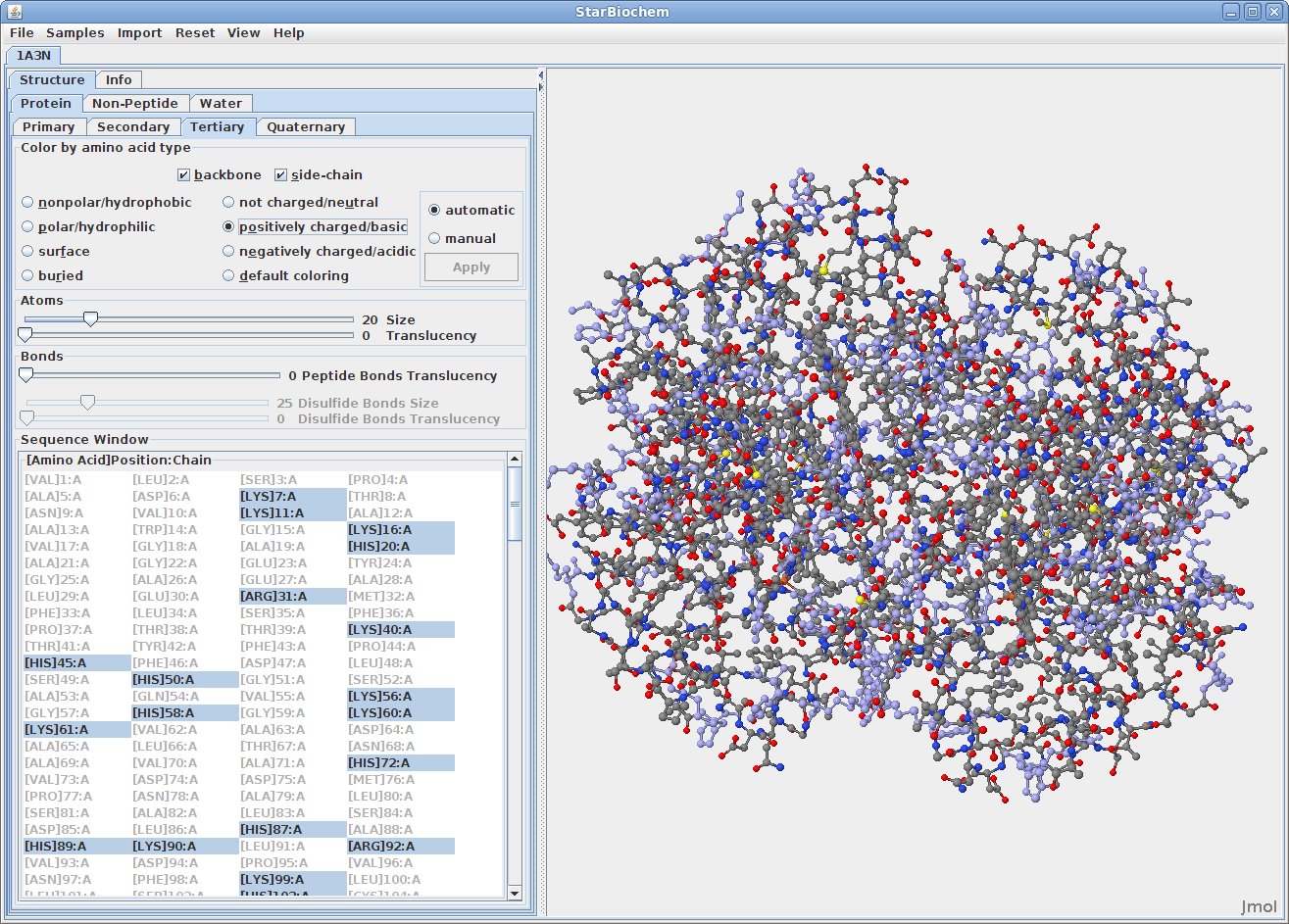
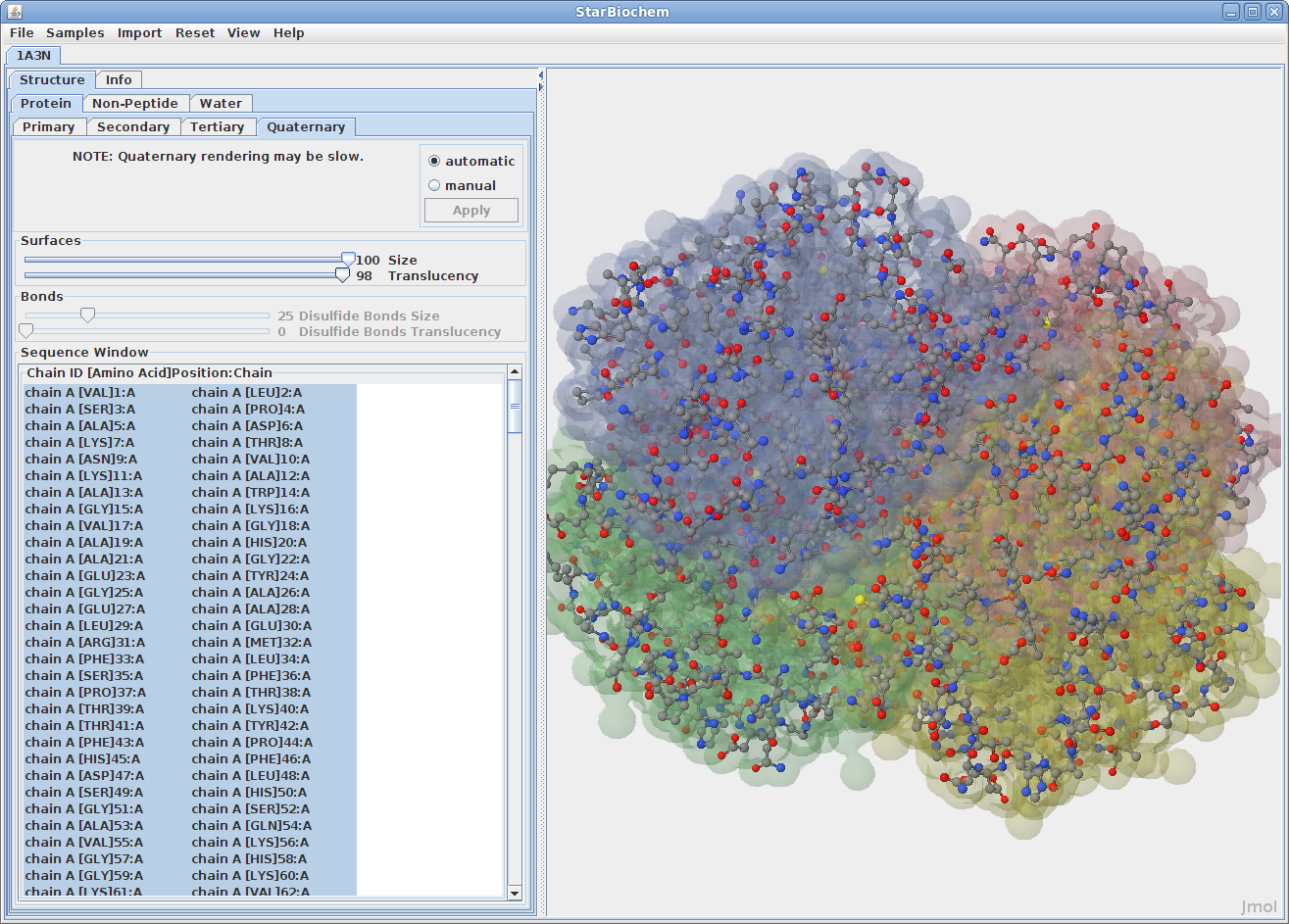
The primary structure of a protein lists the amino acids that make up a protein’s sequence, but does not describe their corresponding shape. The secondary structure of a protein refers to how stretches of amino acids within a protein chain are arranged in characteristic patterns such as helices, sheets, and coils. The tertiary structure describes the folded shape of a protein chain and is determined by characteristic properties of the amino acid residues, such as acidic, basic, polar, and non-polar. The quaternary structure indicates the relationship between the chains of a molecule.
Displaying primary structures:
Displaying secondary structures:
Displaying tertiary structures:
Displaying quaternary structures:
Disulfide Bonds:
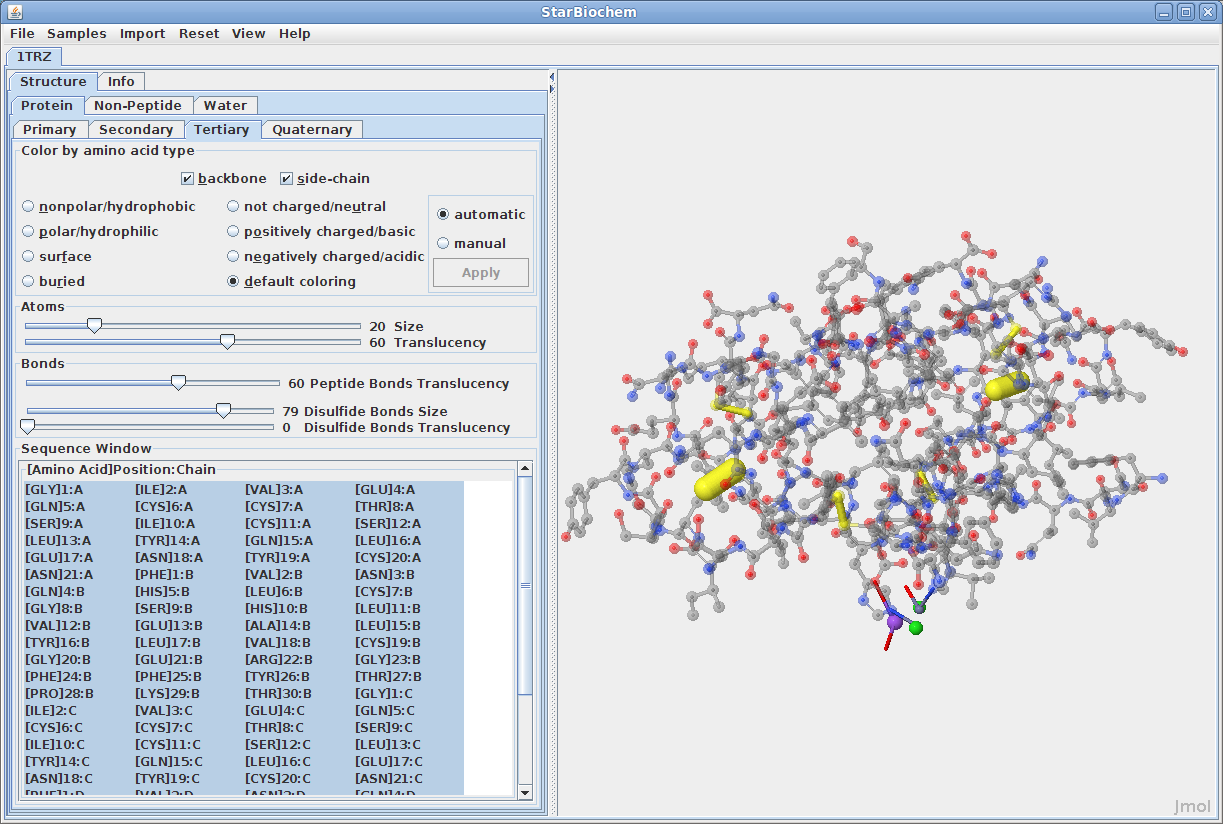
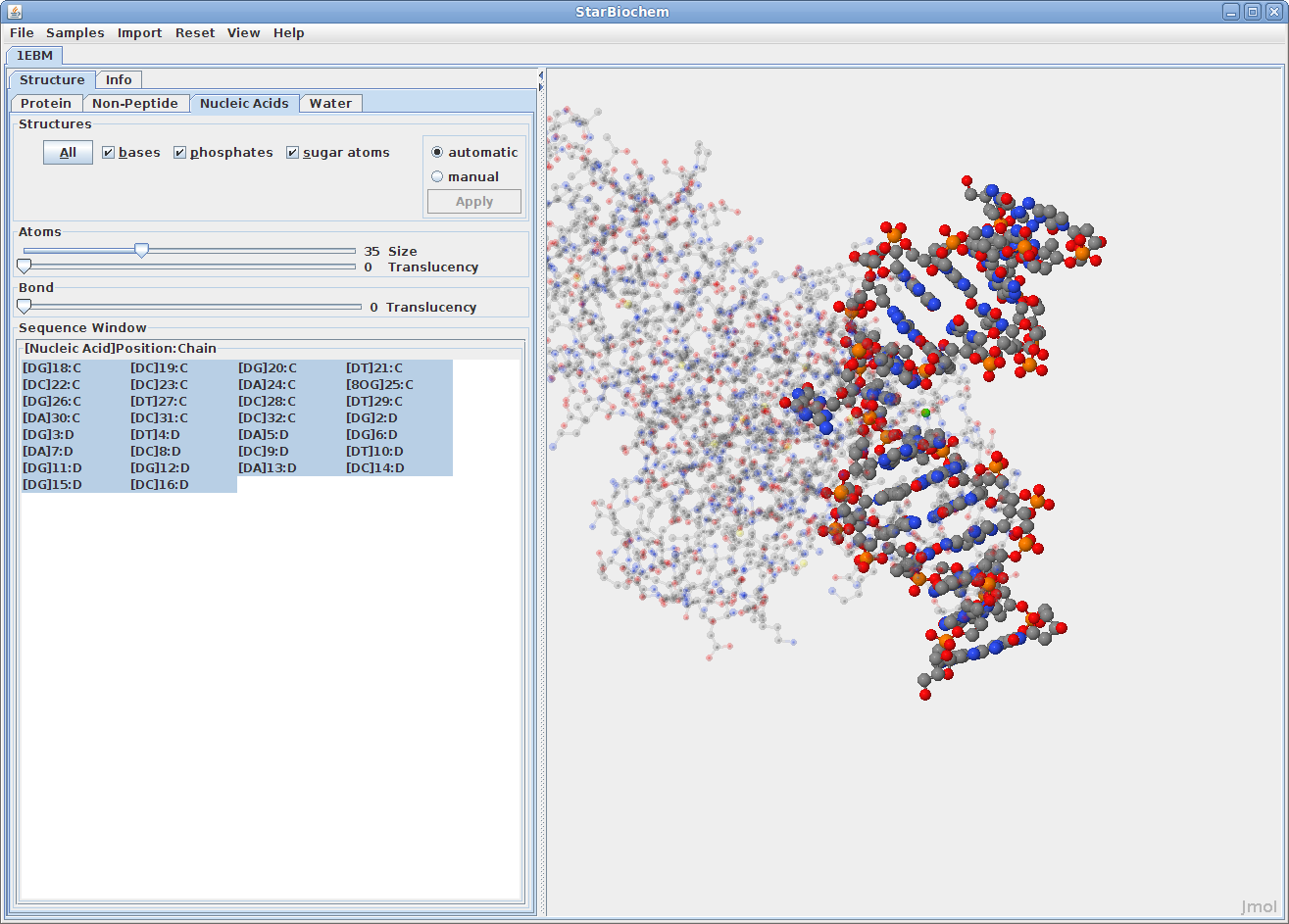
Displaying nucleic acids:
Click on the Nucleic
Acids tab.
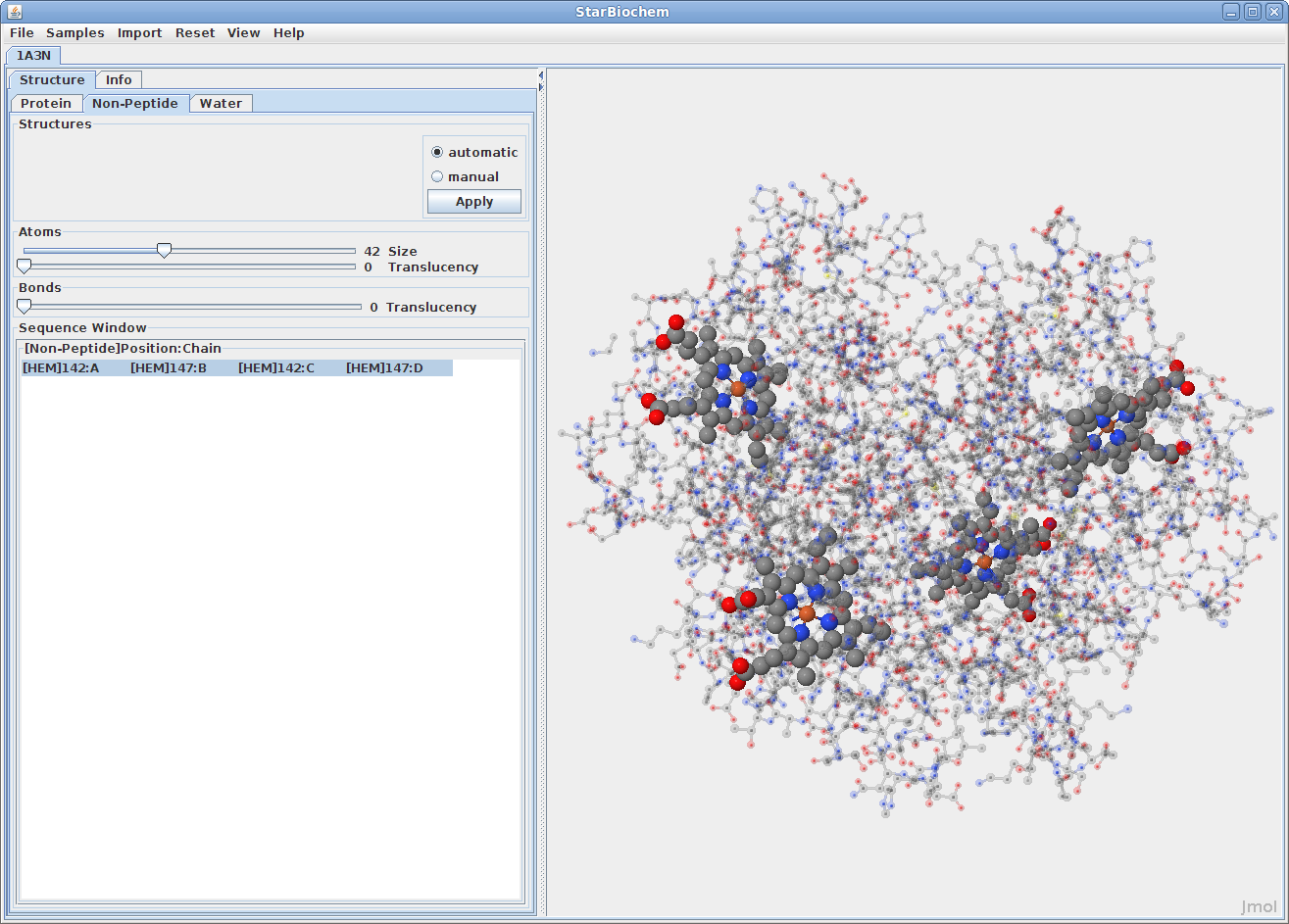
Displaying non-peptides:
Some proteins have the addition of a Non-Peptide tab. Select this
to view any non-peptide atoms within the protein and
change their size in the viewer.
Displaying water atoms:
Many proteins have the addition of a Water tab. Select this tab
to see the oxygens that make up water molecules
within the protein (hydrogens rarely defined).
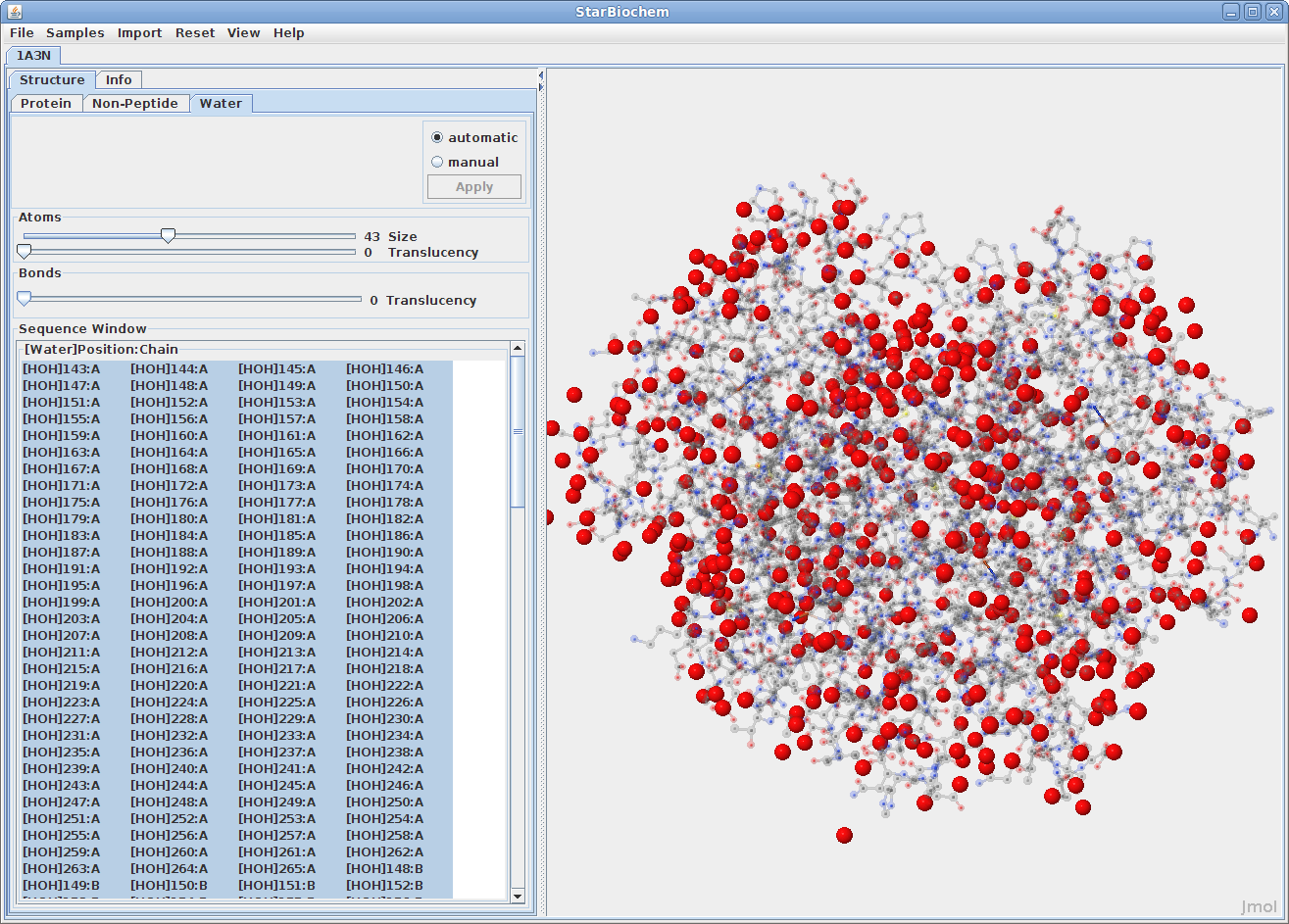
By default the size of the water atoms is set to zero. Move the Size slider to the right to make them visible.
StarBiochem allows for selection of groups or individual atoms and structures.
Selecting Sequences:
Selecting Quaternary Sequences:
StarBiochem has several additional tools to manipulate structures.
Reset:
In the top menu select Reset to view the reset
options.
Show/Hide Hydrogens:
Most pdb files do not show the hydrogen atoms for the
structure.
Viewing specific regions and setting the center of rotation:
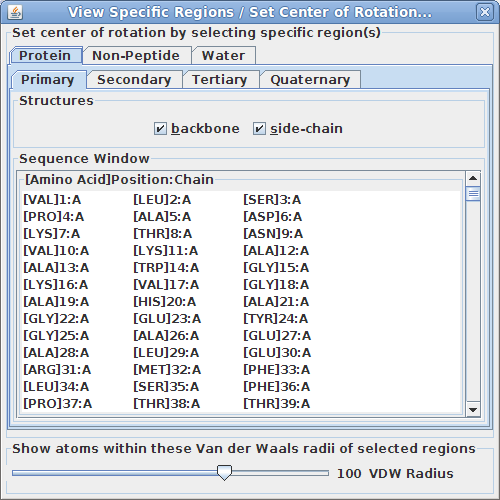
This tool is particularly useful for isolating specific region(s) of the structure to examine more closely how they interact.
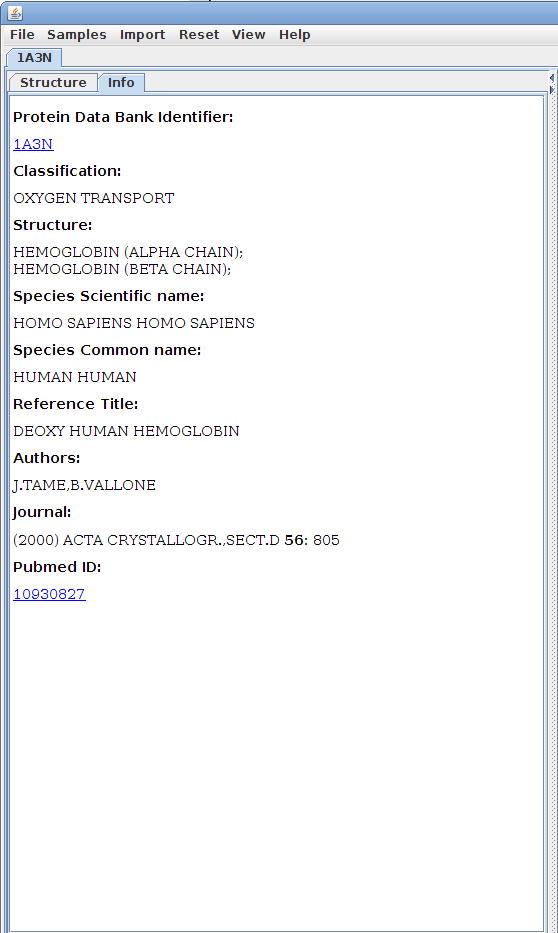
The Info tab:
The Info tab
contains identifying details about the structure and
offers resources for further research.
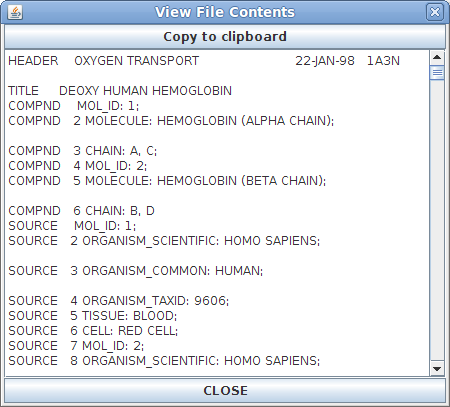
File Contents Window:
The File
Contents window contains the exact contents of
the PDB file that is open. This window is primarily
useful for identifying a damaged PDB file or for
advanced users who are familiar with how PDB files
are written and work.