

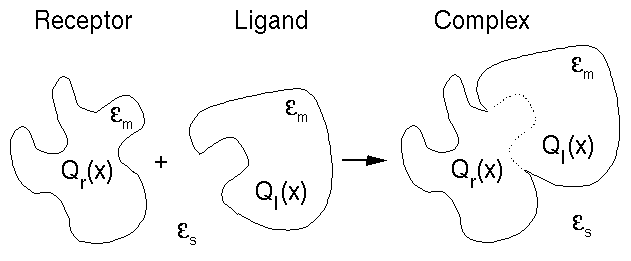
In a binding process such as that shown in Figure 1, two molecules (a ligand and a receptor) associate in solution to form a complex. If each of the three individual molecules exist predominantly in a single conformation, then the free energy change due to the binding process can be separated into electrostatic and non-polar components [1,2]. The electrostatic component of the binding free energy is, in the continuum electrostatic approximation, quadratic [3,5] with respect to the ligand- and receptor-charge distributions. If the electrostatic charge distribution of the ligand is varied while the ligand and receptor shapes and the binding conformation are kept fixed (i.e., all non-polar aspects of binding are held constant), then an optimum set of electrostatic charges can be found for the ligand that make the electrostatic component of the binding free energy as favorable (negative) as possible [3-5].
A ligand whose electrostatic charges are optimized in this way is denoted a complementary ligand because its charges are electrostatically complementary to the given receptor and because it binds to the receptor with the best possible electrostatic free energy change compared to any other ligand of the same shape with differing charges. This optimization process leads to a principle of complementarity [5] between a receptor and its optimal ligand:
This leads to the definition of the Residual Potential:
The Residual potential allows one to rigorously determine and analyze ligand regions that are not complementarity to a given receptor. This can be done explicitly by plotting or contouring the Residual Potential on the ligand's surface to discern regions where it is significantly non-zero, as shown in the section Example of Employing the Residual Potential. With the Residual Potential, one can
Analysis through the Residual Potential is inherently asymmetric because receptor and ligand cannot generally be mutually complementary. As a result, the degree of complementarity observed using the Residual Potential may depend on which of the two associating molecules is labeled the ligand and which the receptor. When actually optimizing the ligand, this choice is unambiguous; however, for natural complexes, it is not. In fact, by considering both possibilities, one may be able to determine which of the associating species is more complementary to the other and thus gain insight into the evolutionary history of the system in question [5].
In the next section, Examples, we show how the Residual potential can be used to analyze complementarity and compare this method to the usual method used by structural biophysicists, which involves only examining the surface electrostatic potentials of the free ligand and receptor. In the section Software, we give scripts for computing and displaying the Residual Potential using the GRASP [6] software package.


![[Tidor Lab]](../images/blue_swirl/labhome_new.jpg)
![[Residual Pot.]](../images/blue_swirl/residual_new.jpg)
![[Examples]](../images/blue_swirl/examples_new.jpg)
