
Research
| Protein conformation and dynamics | ||
|
The folding and association of proteins arise from a complex interplay of non-covalent interactions involving the protein and water with kinetics that span picoseconds to hours. Because of the complexity of these multi-scale structural evolutions, we seek to identify and describe motion in terms of both direct structural coordinates and collective coordinates, to answer general questions about the process by which protein chains find specific contacts:
Our approach has been to use 2D IR spectroscopy of the amide vibrations of the polypeptide backbone. The secondary structural sensitivity of amide I vibrations and the picosecond time resolution of vibrational spectroscopy makes 2D IR an excellent probe of protein dynamics. 2D IR experiments are used in combination with laser temperature jumps to probe protein unfolding and dissociation on nanosecond to millisecond time scales. We complement excitonic amide I spectroscopy with isotope labels to probe site-specific structure and disorder. These experiments are complimented by accurate structure-based models that allow us to calculate spectra from MD simulations. Amide I Two-Dimensional Infrared Spectroscopy of Proteins, Z. Ganim, H.S. Chung, A. Smith, L.P. DeFlores, K.C. Jones and A. Tokmakoff, Acc. Chem. Res. 41(3), 432-441 (2008). |
|
|
| β-Hairpin Folding | ||
|
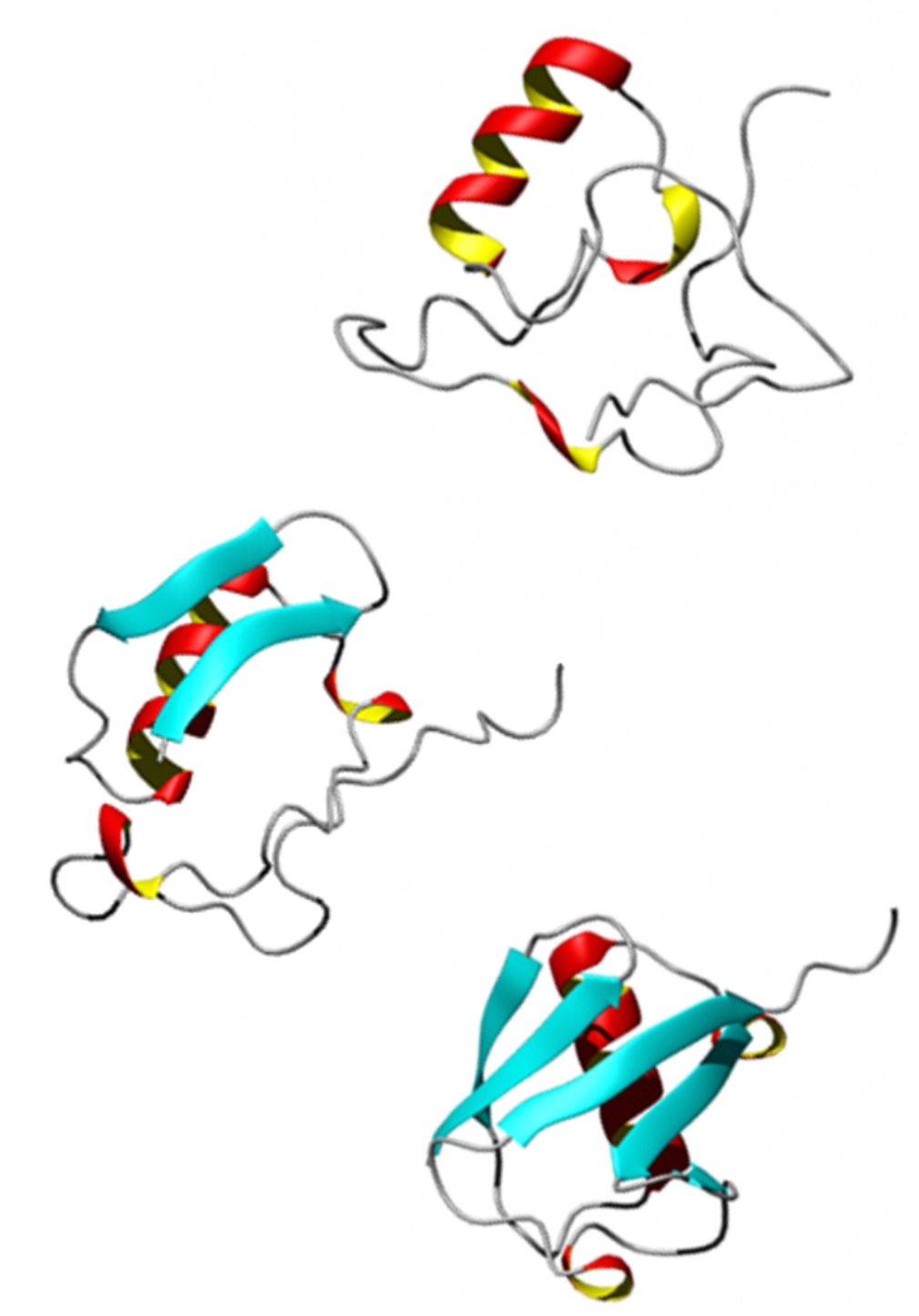
We use β-hairpins as model systems to investigate the role of hydrogen bond contact formation and backbone configurational changes in folding, and to understand the structural heterogeneity of folded states. Hairpins can be synthesized with isotope labels to provide multiple local probes of structure. Our studies of folded TrpZip2 reveal variation in the structure the turn and fraying of ends of the strand, as well as the shifts in population between conformers that occur with increasing temperature. Our T-jump experiments also reveal the nanosecond-microsecond kinetics of unfolding and conformer exchange, suggesting a highly heterogeneous folding process. “Melting of a β-hairpin peptide using isotope-edited 2D IR spectroscopy and simulations,” A. W. Smith, J. Lessing, Z. Ganim, C. Sam Peng, A. Tokmakoff, S. Roy, T. L. C. Jansen, and J. Knoester, Journal of Physical Chemistry B 114, 10913–10924 (2010). Probing local structural events in β-hairpin unfolding with transient nonlinear infrared spectroscopy,” A. W. Smith and A. Tokmakoff, Angewandte Chemie, International Edition 46, 7984- (2007). |
 |
|
| Molecular recognition and binding | ||
|
We use the insulin monomer-to-dimer transition as a model system to investigate conformational changes associated with protein-protein interactions. By designing dynamics experiments, we can understand how monomers encounter and recognize one another, how proper registry is found to yield a specific interaction, and what conformational changes are needed for dimer formation. We combine temperature jump 2D IR spectroscopy with MD simulations of dimer dissociation to reveal the contacts necessary to dimerize, the interplay between secondary structure formation and diffusion, and illuminate the role of desolvation and solvent hydrogen bonding. “Insulin Dimer Dissociation and Unfolding Revealed by Amide I Two-Dimensional Infrared Spectroscopy,” Z. Ganim, K. C. Jones and A. Tokmakoff, Physical Chemistry Chemical Physics 12, 3579–3588 (2010) . |
 |
|
| Conformation and folding of elastin-like peptides | ||
|
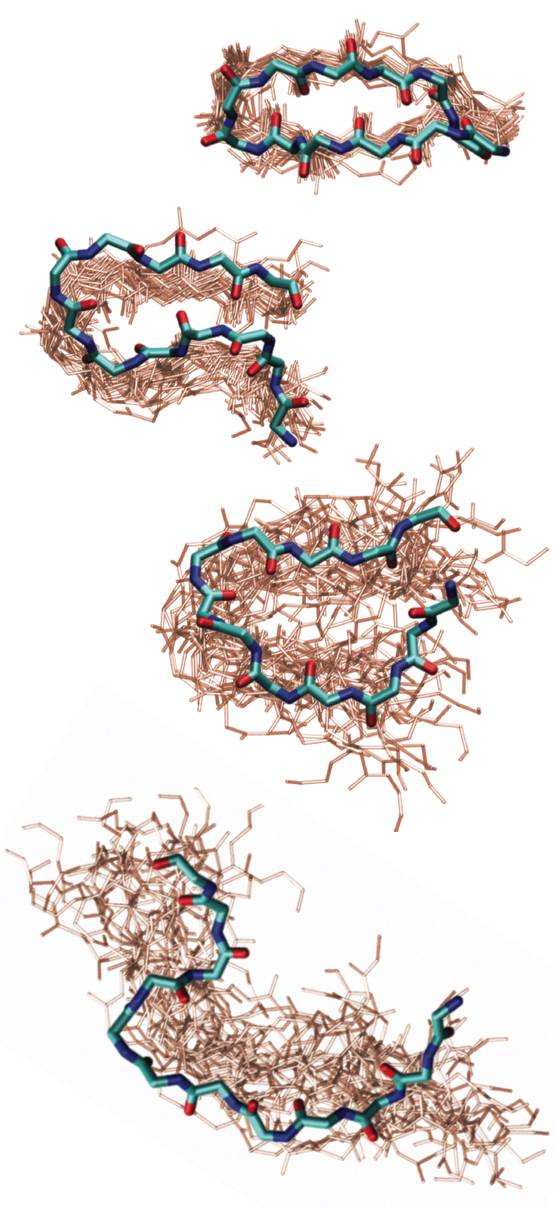
Elastin is the protein of our connective tissue that gives its elastic properties. The unusual extensibility and tensile strength of elastomeric proteins have been related to amino acid repeat sequences of the form (XPGXX)n, where X are ususally hydrophobic side-chains. Many structural proposals have been made for these chains, focusing on the turn propensity of PG, and it has been postulated that hydrophobic hydration is the origin of their restoring force. We are investigating the structure and folding of elastin-like peptides of the sequence (VPGVG)n in order to understand the physical origins of the structure and conformational changes on extension and contraction. “Identifying residual structure in intrinsically disordered systems: A 2D IR spectroscopic study of the GVGXPGVG peptide”, J. Lessing, S. Roy, M. Reppert, M. Baer, D. Marx, T. La Cour Jansen, J. Knoester, and A. Tokmakoff, J. Am. Chem. Soc. 134, 5032-5035 (2012). |
 |
|
| Protein-water interactions | ||
|
Our current method developments are aimed at developing insight into the interactions between protein and water. One example of this work is our development of multimode 2D IR spectroscopy of the Amide I and II vibrations. This technique uses 2D IR in conjunction with H/D exchange measurements to correlate the secondary structure sensitivity of amide I vibrations with the sensitivity of amide II to solvent exposure. In this experiment a protonated protein is placed in deuterated water which induces H/D exchange of amide protons. As a result of deuteration, the Amide II vibration red shifts by 100 cm-1 (Amide II′). By observing the 2D IR cross peak of Amide II and II′ with Amide I it is possible to map information on water accessibility in the protein. “Water penetration into protein secondary structure revealed by hydrogen-deuterium exchange 2D IR spectroscopy,” L. P. DeFlores and A. Tokmakoff, Journal of the American Chemical Society 128, 16520-16521 (2006). | 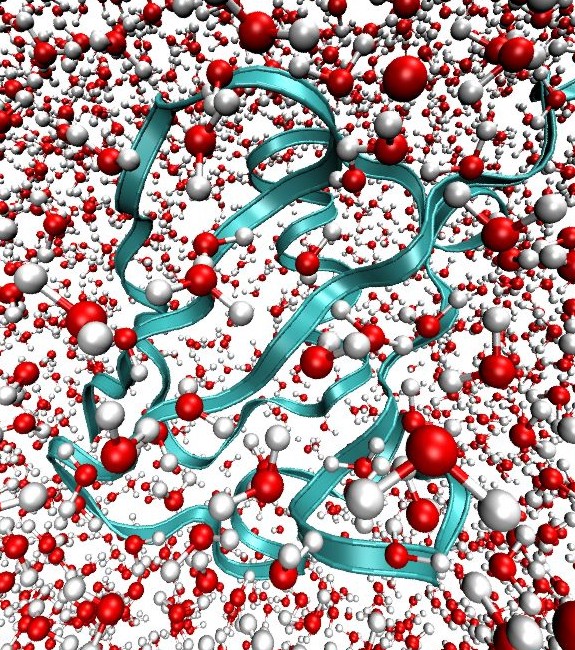 |
|
| Ion Transport in Membrane Proteins | ||
|
Membrane proteins are key components to a number of biological functions ranging from homostasis to signaling; however, traditional techniques for studying protein structure are perturbative or poorly suited to membrane proteins. We are developing 2D IR spectroscopy as a tool to probe the secondary and tertiary structure of membrane proteins in situ. This work involves methods to obtain large scale structure and methods to study ion transport by the KcsA potassium channel. |
 |
|