Research
Overview
How do genes control animal development and behavior? To answer this question, we use the experimentally tractable nematode Caenorhabditis elegans to identify and analyze molecular and cellular pathways involved in these important areas of biology, with the goal of discovering fundamental biological mechanisms and revealing insights into human diseases.
Research Summary
Programmed Cell Death
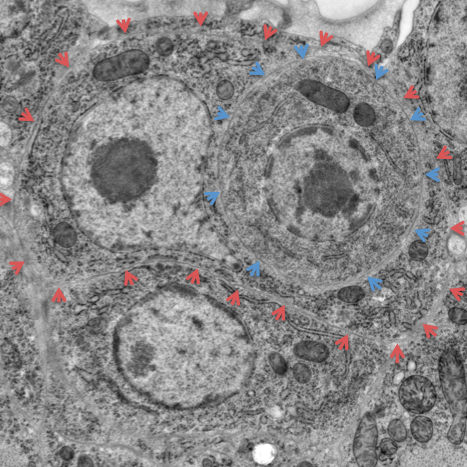
Naturally-occurring or “programmed” cell deaths play major roles in animal development and homeostasis, and abnormalities in programmed cell deaths are causally involved in numerous human diseases, including cancer and certain neurodegenerative disorders. Diverse developmental events involve programmed cell deaths. The loss of the tadpole tail during metamorphosis into a frog, the sculpting of human fingers and toes in utero, the seasonal degeneration of the brain region that drives birdsong, and the elimination of self-reactive cells in the immune system all provide examples of programmed cell death. Cells that die by programmed cell death undergo a process known as apoptosis, defined by morphological and ultrastructural characteristics. Abnormalities in programmed cell death are causally involved in numerous human diseases, including certain neurodegenerative disorders and cancer. We discovered that the 131 programmed cell deaths that occur during C. elegans development are controlled by a specific set of killer and protector genes, and we defined the canonical molecular genetic pathway for programmed cell death. This pathway of cellular suicide subsequently proved to be conserved in other animals, including humans, and the human homologs of genes we discovered are now therapeutic targets for human diseases. One of the C. elegans killer genes, ced-3, encodes the founding member of a family of proteases known as caspases, key drivers of apoptotic cell death. We recently discovered that in C. elegans some cells die in the absence of caspases. We currently are exploring a number of major problems in the field of cell death:
- how specific cells decide whether to live or die
- how caspase activation kills cells
- how neighboring cells recognize and engulf dying cells to eliminate them
- mechanistic differences among cell suicides, murders, induced suicides and assisted suicides
- mechanisms responsible for caspase-independent apoptosis
- how caspase-dependent and caspase-independent cell-death pathways are coordinated within single cells
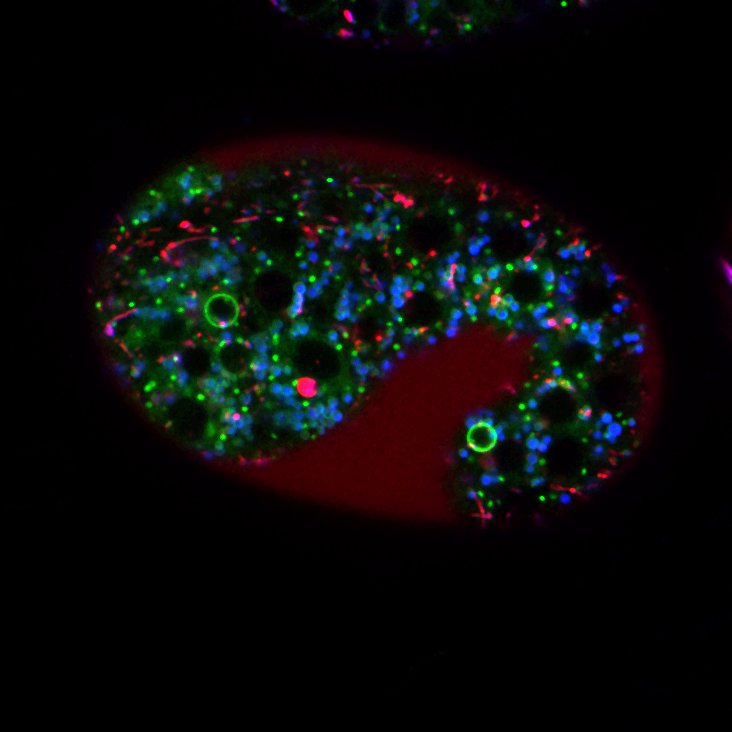
Cell Lineage and Cell Fate
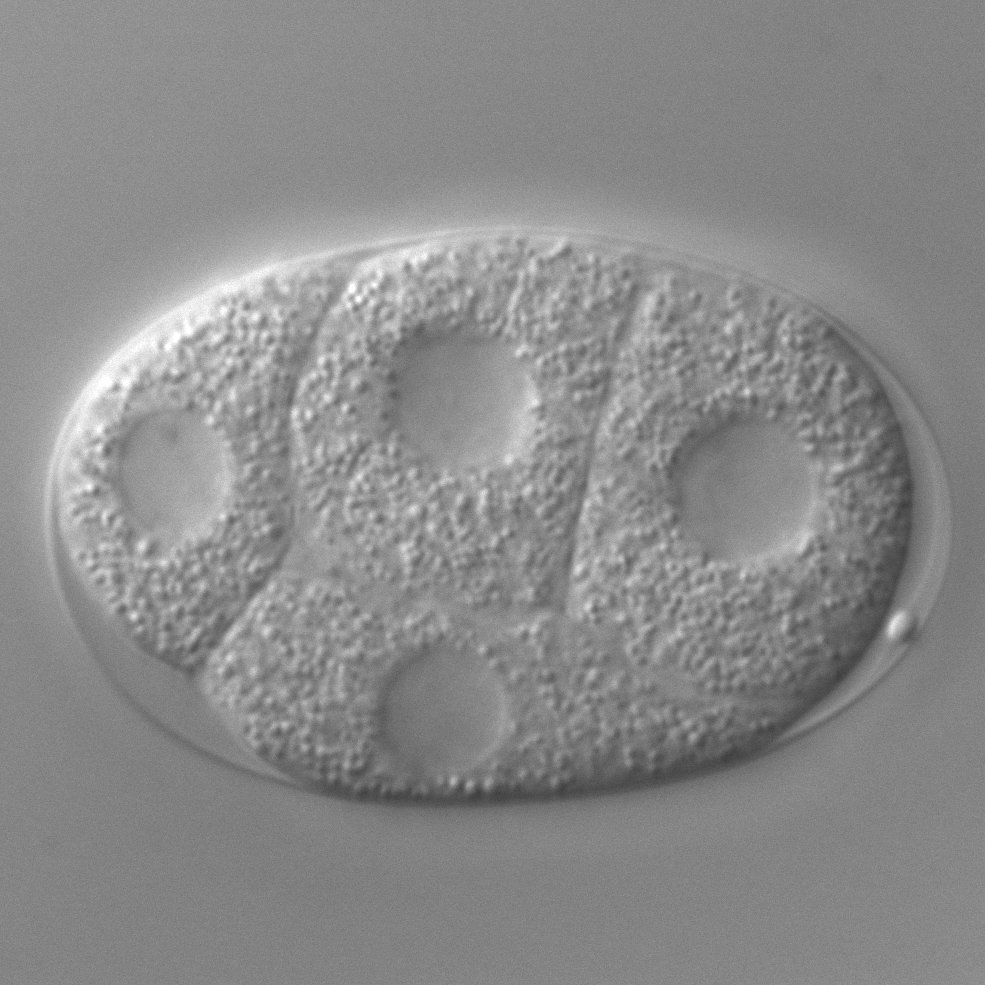
Animal development begins with the fertilized egg and proceeds through many cell divisions to generate a great variety of different cell types. How cell diversity is generated during development is a fundamental issue in biology. C. elegans has only 959 somatic cells and a known cell lineage, facilitating analyses of development at the single-cell level. We have identified mechanisms that make daughter cells different from mother cells, that make sister cells different from each other or that break left-right developmental symmetry. Our studies of the induction of vulval development helped define the Ras signal transduction pathway, key in cancer biology. We defined the heterochronic pathway, which controls developmental timing and led to the discovery of microRNAs. We currently are analyzing the following issues involving cell lineage and cell fate:
- how neurons can be generated from mesodermal cell lineages
- mechanisms that control the developmental timing of the expression of cell fates
- how epigenetic factors determine cell fates
- mechanisms that control the maintenance, as opposed to the establishment, of cell fates
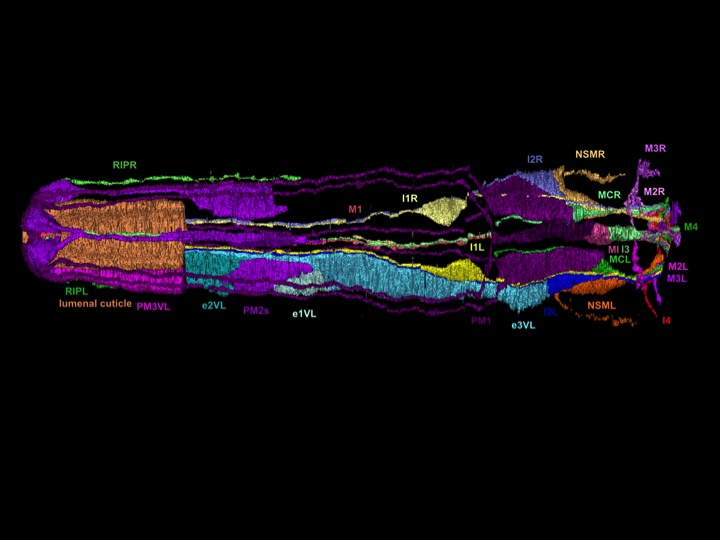
Behavior
To explore the molecular, cellular and circuit bases of nervous system function, we are analyzing the coordinately controlled C. elegans behaviors of feeding, egg laying and locomotion. From our studies of behavior we have identified founding members of the RGS protein family, which regulates signaling from G-protein coupled receptors, and the EGLN family of dioxygenases, which control the HIF-VHL pathway that is vital in cellular and organismic O2 sensing and is a therapeutic target in cancer. We also identified a novel class of neurotransmitter receptors, chloride channels gated by biogenic amines, and a new neurotransmitter, tyramine. Our current studies include the following problems:
- how the EGL-9/HIF pathway, key in cancer biology, controls animal behavior in response to changes in O2 concentration
- how neuropeptides and biogenic amines modulate animal behavior and behavioral state
- how neural regulation in response to noxious stimuli convert swallowing to spitting behavior
- how maternal stress drives signaling from the nervous system to the germline to confer stress-resistance to progeny
- mechanisms that control muscle metabolic homeostasis in response to changing environmental conditions
- mechanisms that function in classical conditioning
- how hyperglycemia drives neuropathy
Amyotrophic lateral sclerosis (ALS)
We have long studied the human neurodegenerative disease ALS. In collaborative studies, we have identified and characterized many ALS genes, including the first known ALS gene, SOD-1. We are now analyzing:
- the function of the C. elegans ortholog of the human gene C9orf72, in which hexanucleotide-repeat expansion disease alleles cause both ALS and frontotemporal dementia.