|
1) Answer: A, H, I, N Genes are composed of stretches of DNA that code for proteins (exons) interspersed with intervening DNA sequences that do not (introns). At the onset of transcription, genomic DNA is copied (transcribed) into early mRNA. In a series of steps, collectively known as splicing the introns are removed (spliced out) yielding mature mRNA with one continuous protein coding region (the open reading frame), flanked by sequences that direct protein expression and mRNA localization within the cell (the untranslated regions or 'UTRs').This is a typical Gene Map. It demonstrates the arrangements of introns and exons in the genomic DNA and also indicates which stretches of DNA encode protein. Further details can be found in the Gene Structure, Transcription and Splicing sections of the Resource Guide.
2) Answer: B, H, J, M This question covers the process of transcript modification, which sees mRNA from its early form (as it has just been copied from the DNA) to its final form wherein it serves as a template for protein synthesis. Further details can be found in the Transcription and Transcript Modification sections of the Resource Guide.
3) Answer: D - 1886 and 1075 Digestion of circular DNA with an enzyme (or enzymes) that cuts n times in the plasmid will yield n fragments. In this case, each enzyme cuts once thus two fragments will be produced. The KpnI-XmnI fragment will be 2645-759=1886 bp, while the size of the remaining fragment can be determined by subtracting this value from the total size of the plasmid (2961 bp) yielding a fragment of 1075. Further details can be found in the Restriction Endonucleases section of the Resource Guide.
4) Answer: C - 9.7, 4.9, 3.1, 2.0 and 0.6 Here is a breakdown of the fragments produced reading the figure from left to right:
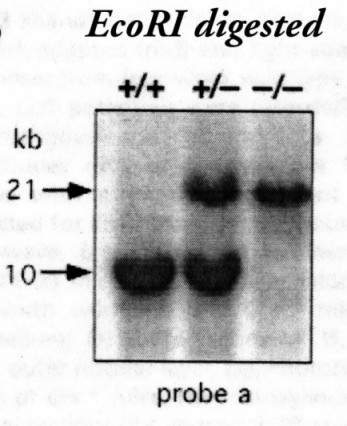
5) Answer: D - 10.3 and 19.3 This type of analysis can be tricky inasmuch as gene maps tend to contain far more information than is really need to get your answer. The simplest approach is to generate a list of DNA fragments that will result from digestion of your DNA with a given enzyme. In this case, digestion of wild type genomic DNA with EcoRI will generate fragments of 6.9, 3.1 and 10.3 kb, while digestion of the knockout will generate a single fragments of 19.3 bp. The band or bands that will appear on a Southern blot then correspond to the fragment(s) of DNA overlapped by the probe. In this case, the probe falls entirely within a single EcoRI fragment, thus only a single band will appear after hybridization in either background. Here is an autoradiogram of a Southern blot used to distinguish Crx wild type, heterozygous and homozygous knockout mice, probed with 'Probe A' (the DNA size markers labelled on the left are only approximations).
As practice, you may want to repeat this exercise determining the sizes of bands on a Southern blot of DNA digested with BamHI and hybridized with probe B. Click here for the answer (this will open a new window). For a further description of blotting methodology and figures, refer to the Blotting section in the resource guide.
6) Answer: B - open reading frame This is the open reading frame from a gene and represents the protein-coding sequence. It is formed once the introns have been spliced out of the mRNA, thus apposing the exons. The codons are the three-base sequences that indicate which amino acid is to be added to the polypeptide chain. For any coding sequence of length n, the resulting protein will have n/3 amino acid residues. For a further discussion of these principles, refer to the Gene Structure, Transcription and Processing and Translation and Protein Structure sections in the resource guide.
7) Answer: B - 505 Every amino acid of a protein is encoded by one codon. Each codon is made up of three nucelotide bases. Therefore the number of amino acides in a protein equals the number of bases divided by 3.
8) Answer: D - reverse transcriptase RNA polymerase is used to produce RNA from a DNA template (during transcription). DNA polymerase will produce DNA strands from a DNA template (eg. during DNA replication). A restriction endonuclease digests double-stranded DNA at specific sequences while superoxide dismutase is involved in the processing of free radicals (you will learn more about these in the Pathology and Biochemistry courses). For more information on these topics, consult the Central Dogma and Recombinant DNA and Cloning sections of the resource guide.
9) Answer: C - I and VI For PCR, one requires a 'forward' and a 'reverse' primer. The forward primer will have a sequence identical to the that at the start of the portion you are trying to amplify. The 'reverse' primer will be the 'reverse complement' of the sequence at the end of the DNA that you are amplifying. For more information, see the PCR section of the resource guide.
10) Answer: B - nonsense This mutation converts a GGG codon (which codes for Glycine) to UGA which is a stop codon. This will introduce a premature termination and thus the nascent polypeptide will be shorter than normal. By definition, a "nonsense" mutation is a mutation that introduces a premature stop codon. In contrast, a "missense" mutation will change the amino acid coded for at that codon but will not alter the polypeptide length (ie. it may change an Alanine to a Glycine residue). A "silent" mutation has no effect on the sequence of a polypeptide, while a "frameshift" mutation results from the insertion or removal of nucleotides in the DNA (the number of nucleotides inserted or removed will not be divisible by 3). It alters the reading frame and thus changes both the amino acid sequence following the mutation and, most often the polypeptide length. The concept of a frameshift mutation is conceptually the most difficult, but it is essential that you understand it. For a further description of translation, refer to the Translation and Protein Structure and Mutations sections in the resource guide.
11) Answer: B - shorter The nonsense mutation introduces a premature stop codon and thus a shorter protein
12) Answer: C - silent In this situation, the codon GGA has been changed to GGC. Both of these codons specify glycine and thus there is no effect on the polypeptide produced.
13) Answer: B - reduced signal in Western blot only If one suspects that the level of the protein produced is decreased, but the level of the RNA is unaffected, then then bands on a Western blot will be reduced in intensity, while the intensity of the bands on the Northern blot will be unchanged. This situation is often referred to as a post-transcriptional effect. In other words, transcription (of RNA) is normal but following this event, something causes the amount of protein produced to be affected. Blots of RNA are known as Northern Blots, while blots of DNA are known as Southern blots. It is important to recognize that if RNA levels were decreased (as seen on a Northern blot), it is very likely that protein levels would also be decreased. For more information, see the Blotting section of the resource guide.
14) Answer: D - histones Histones are a complex of proteins around which DNA strands are wrapped and thus compacted. Acetylation of histones is a key event in the "unwrapping" of genomic (or chromosomal) DNA in preparation for transcription. Acetylation is a reversible modification of the proteins and is mediated by a protein known as Histone Acetyl Transferase (HAT). To reverse the process, a protein called Histone Deacetylase (HDAC) removes the acetyl groups from histones, thus closing that region of DNA to transcription. For more information, see the Packing into Chromosomes section of the resource guide.
15) Answer: D - acetylation Please see the answer to Question 14.
16) Answer: A - promoter The promoter is the site of binding for RNA polymerase along with a large number of other proteins and it is always upstream (5') of the transcriptional (and translational) start site. The enhancer modifies the level of gene expression, but may be located upstream, downstream or in the middle of a gene. See the section on Control of Gene Expression for more details.
17) Answer: A - upstream Please see answer to Question 16.
18) Answer: B - protein => DNA The central dogma states that DNA is used as a template for RNA (transcription) which is, in turn, used as a template for protein synthesis (translation). RNA may also be converted back to DNA by 'reverse transcriptase'. While proteins are involved in the replication of DNA, they never serve as templates for the synthesis of DNA. See the Central Dogma section of the resource guide for more information.
19) Answer: D - 5'-CAGTCTAGTCCCGACTCGATC-3' To determine the sequence of a complementary strand of DNA, it is important to recall that complementary DNA strands pair up in an anti-parallel glossary manner. This means that the 5' end of the complementary (antisense) strand will line up with the 3' end of the original (sense) strand. Since the 3' end of the sense strand reads GTCA (reading 3' to 5'), the complementary strand must begin (5' to 3') CAGT. Only answer D fits this requirement. See the Base Pairing section of the resource guide for more information.
20) Answer: B - 4 From the information given in the question, one can assume that blue eyes are a recessive trait. As such, AG must be homozygous recessive for the allele, while we can assume that TG is homozygous (dominant) for brown eyes. In this situation, all of their offspring will be heterozygous for the two alleles and if each of their spouses are also heterozygous, 1/4 of AG and TGs grandchildren will be homozygous recessive. From a total of 18, 1/4 would be 4.5 so we can assume that 4 is the closest answer. Ok, the question is a bit far-fetched but, you get the picture! For further information, see the section on Mendelian Genetics in the resource guide.
21) Answer: A - 1 Using the same logic as question 16, independently each recessive trait will be inherited by 1/4 of the grandchildren. The probability of being doubly homozygous for the recessive trait is thus the product of the probabilities for being homozygous for each trait. Therefore, double homozygous recessive grandchildren can be expected to appear at a frequency of 1 in 16. In this situation 1 would be the closest answer. Note that the same odds would apply to doubly homozygous dominant, however we can't distinguish these individuals from heterozygotes by phenotype (homozygous dominant and heterozygous individuals will all have brown hair and brown eyes). For further information, see the section on Mendelian Genetics in the resource guide.
22) Answer: B - meiosis Recombination (or 'crossing over') occurs during meiosis and is a key event in establishing diversity of the gene pool Meiosis is a sole feature of the germ cells (both male and female). So-called "somatic recombination" occurs at a very low level in any cell in an organism. It can be the predisposing event in the transformation of cells towards a cancerous fate. More information can be found in the Recombination section of the resource guide.
23) Answer: C - germ cells Please see the answer to Question 22.
24) Answer: D - both a missing chromosome and an extra chromosome Non-disjunction describes the failure of homologous chromosomes to segregate into each of two daughter cells. This will then leave one cell without said chromosome, while the other daughter cell with an extra. Trisomies (for example Trisomy 21 which is associated with Downs Syndrome) result from non-disjunction.
25) Answer: B - isogenic Isogenic animals are incredibly valuable tools for laboratory research as they allow the researcher to compare the effects of changes at a single genetic locus in animals of an otherwise identical genetic background. You will hear more about this when we discuss animal models of disease. The glossaries in the resource guide contain definitions of common genetic terms.
26) Answer: B - maternal pattern Mitochondrial genes are always inherited in a maternal fashion. Mitochondrial DNA is a very useful tool in DNA-based identification. Two entertaining exercises in the Mitochondria and Mitochondrial DNA section of the resource guide can be used to gain more insight into this aspect of genetics.
27) Answer: B - autosomal recessive Since menin is located on Chromosome 11, it must be an autosomal trait. Also, given that only 25% of the protein is required for normal function, mutation of one copy of the gene would most likely not have a substantial deleterious effect, As such, we can guess that MEN type I is inherited as an autosomal recessive trait. You should note, however, that MEN type I often occurs in individuals who are heterozygous at the menin locus (and thus phenotypically normal) but then undergo a somatic mutation causing inactivation of the second allele, thus setting the disease in motion. Consult the section on Mendelian Genetics in the resource guide for further information on this topic.
28) Answer: A - autosomal dominant This type of mutation is common among oncogenes. A given protein involved in a regulatory process acquires a mutation which causes it to be constitutively active. In this case, the mutation is dominant (only one copy needs to be mutated in order to produce the phenotype). In fact, two different activating mutations in ret have been identified and associated with MEN type II. Since ret is on Chromosome 10, MEN type II is inherited an autosomal dominant fashion. Consult the section on Mendelian Genetics in the resource guide for further information on this topic.
29) Answer: D - x-linked recessive ALD is an X-linked recessive disorder. You'll notice that roughly 50% of sons of mothers carrying the mutation are affected, while none of their female siblings are. Consult the section on Mendelian Genetics in the resource guide for further information on this topic.
30) Answer: B - The patient initially found to carry a genetic defect, around which linkage analysis is performed It is important that you become familiar with commonly used genetic terminology. The two Glossaries in the resource guide should help you along the way, but feel free to ask questions if there are any terms that require clarification.
31) Answer: D - HSTGENETICS (boy, aren't we clever) To 'translate' a DNA sequence, simply divide the sequence into three-base long stretches and determine which amino acid is specified by each codon. The genetic code is the "Rosetta Stone" of molecular biology and specifies the translation of each codon. See the section on Translation and Protein Structure for further details.
32) Answer: B - 5'-TGC TAC TTC CAG AAC TGC CCG AGG GGC-3' A polypeptide chain may be encoded by a variety of different DNA sequences. Each amino acid can be coded for by between two and six different codons. This phenomenon is referred to as the 'degeneracy' of the genetic code. Any combination of codons that specify the correct amino acids in their proper order would be sufficient to encode a given polypepetide. Also notice that in this question, the cDNA sequence is required, As DNA contains thymidine (T) rather than uracil (U), you can immediately rule out answers A and D. Of the remaining solutions, B is the correct one.
33) Answer: A - 1 and 6 Disulfide bonds form only between cysteine residues and are important in determining the secondary, tertiary and quaternary structures of proteins. As such, only answer A could be correct. This link provides a graphic illustration of the different classifications of Protein Structure.
|