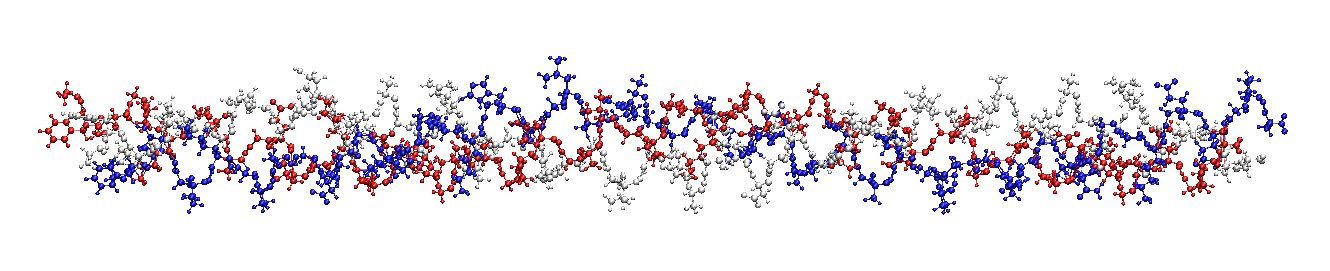


|
from matriarch import *
from math import *
def collagen(seq1, seq2):
a1 = chain(seq1)
a2 = chain(seq2)
hel1 = helixBuilder(a1,1.5,9.5238,'L')
hel2 = helixBuilder(a2,1.5,9.5238,'L')
helhel1 = helixBuilder(hel1,4,85.5,'L')
helhel2 = helixBuilder(hel2,4,85.5,'L')
helhel1rot = shiftOrbs(rotateOrbs(helhel1,2*pi/3),2.8)
helhel2rot = shiftOrbs(rotateOrbs(helhel2,4*pi/3),5.6)
homodimer = overlay(helhel1,helhel1rot)
output = overlay(homodimer,helhel2rot)
return output
def helixBuilder(myBB,rad,pitch,handed):
scale = sqrt(rad*rad + pitch*pitch/(4*pi*pi))
if handed=='R':
sign=1
elif handed=='L':
sign=-1
else:
print handed,' should be L or R.'
def parameterizedHelix(t):
sc = sign/scale
return [rad*cos(sc*t),-rad*sin(sc*t),pitch*t/(2*pi*scale)]
W = buildAxisTwister(parameterizedHelix)
return twist(myBB, W)
seq1 = 'GFZGPKGTAGEZGKAGERGVZGPZGAVGPAGKDGEAGAQGAZGPAGPAGERGEQGPA'
seq2 = 'GFZGPKGPSGDZGKZGEKGHPGLAGARGAZGPDGNNGAQGPZGPQGVQGGKGEQGPA'
collgn = collagen(seq1,seq2)
fileOut(collgn,'collgn.pdb')
|
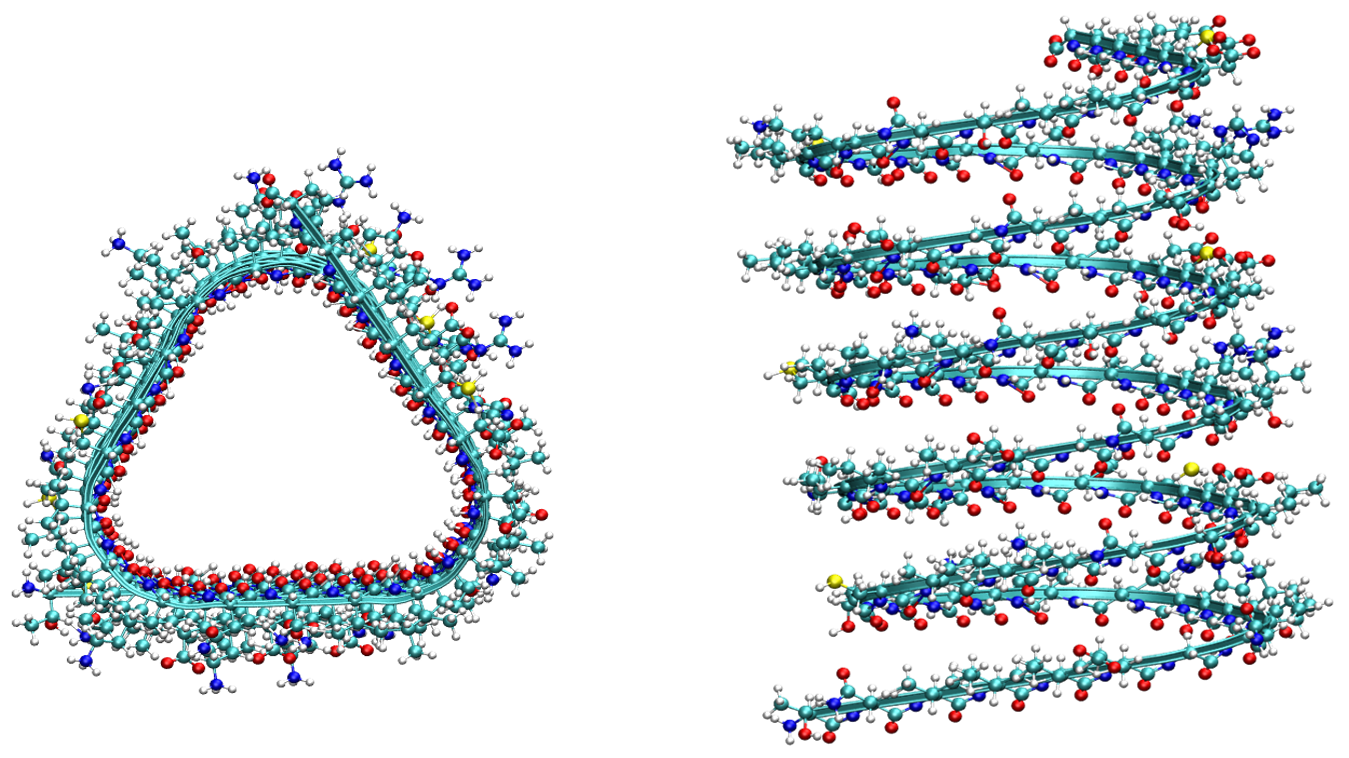
|
from matriarch import *
from math import *
def triangleTwister(side, pitch, length, smoothingFactor):
iterations = int(length/(3*side)) + 2
const = sqrt(3)/6
def loop(Z0):
return [[side*2*const,0,Z0],[-side*const,side/2.0,Z0+pitch/3.0],[-side*const,-side/2.0,Z0+pitch*2/3.0]]
PList = []
for n in range(0,iterations):
PList.extend(loop(pitch*n))
Rout = Ray([0,0,0],[0,0,1])
provideTheta = []
return smoothedPieceWiseLinear(PList, Rout, provideTheta, smoothingFactor)
aminoLength=3.4
actualSeq ='TNVIIEGNVTLGHRVKIGTGCVIKNSVIGDDCEISP'
Mult=3
Side=9*aminoLength
Pitch=7
SmthFact = 0.33
totalLen=Mult*aminoLength*len(actualSeq)
myChain = attachSeries(chain(actualSeq),Mult)
myTriangle = twist(myChain, triangleTwister(Side, Pitch, totalLen, SmthFact))
fileOut(myTriangle,'triangleHelix.pdb')

|
from matriarch import *
from math import *
seq1 = 'AAAAGGPGGYGGPGGAAAA'
a = chain(seq1)
ser = attachSeries(a,25)
def SphSprl(k,Rout):
def curve(t):
return [k*sin(t)*cos(20*t), k*sin(t)*sin(20*t), k*cos(t)]
tmax=pi
Thetaspec=0
return buildAxisTwister(curve,Rout,Thetaspec,tmax)
Rout = Ray([0,0,0], [0,0,0])
k1 = 1 #first try
W1 = SphSprl(k1,Rout)
contourLength = length(ser)
lengthOfCurveWithKEquals1 = W1[0].length
knew = contourLength / lengthOfCurveWithKEquals1
SphSprlTwister = SphSprl(knew + 0.001,Rout)
output = twist(ser, SphSprlTwister)
fileOut(output,'sphericalSpiral.pdb')
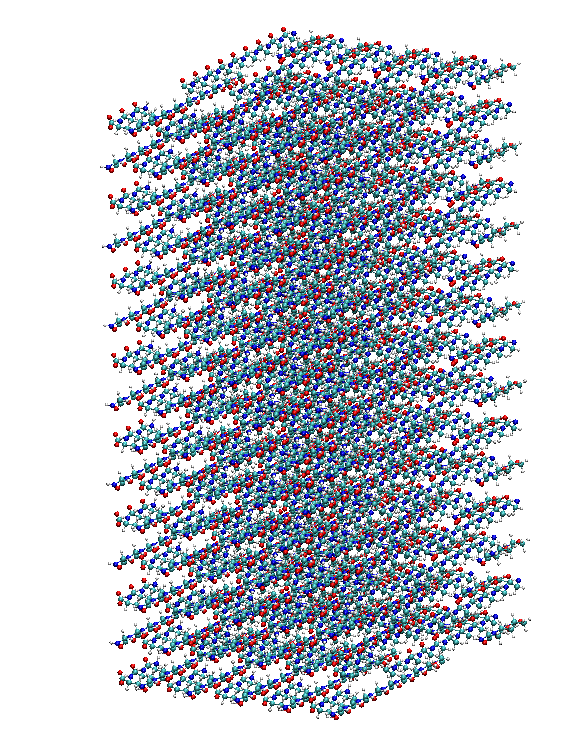
|
from matriarch import * mySeqG = 'GGGGGGGGG' mySeqA = 'AAA' myChainG = chain(mySeqG) myChainA = chain(mySeqA) spacedBlock = space(myChainG, myChainA, 10) array = makeArray(spacedBlock, 10, 15, 4, 6, True) fileOut(array, 'arraySpaced.pdb')
