Research
Cancer Models (primary, metastatic, immuno-oncology)
Vascular Models (lymphatic, BBB, HUVEC+FB)
Neurological Disease Models (Alzheimer's, neuromuscular junction model used to study ALS)
Cancer Models (primary, metastatic, immuno-oncology)
 |
Technologies have been developed to image and study from a mechanistic viewpoint the processes used by tumor cells to form a metastatic tumor. In addition, primary tumor organoids can also be used in in vitro experiments to screen for optimal therapeutic strategies and methods of drug delivery. Our lab has developed model systems designed to produce new understanding of these critical biological phenomena and tools for discovering new therapeutic targets and for personalized medicine. |
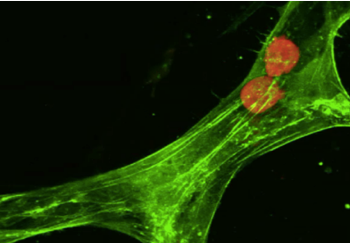
| Model for tumor cell extravasation. Two cancer cells (in red) are trapped inside a segment of a microvascular network (in green). In this time-lapse movie, one of the cancer cells escapes from the vessel through a break in the cell-cell junctions between two endothelial cells. | |
| References: | |
|
|
Vascular Models (lymphatic, BBB, HUVEC+FB)
|
One of the major obstacles to producing realistic microphysiological models of reasonable scale and with long-term viability is the need for a vascular system to delivery nutrients and oxygen to the tissues. Our group has developed microfluidic platforms for a variety of applications including models of metastatic tumors and models or transport across the blood-brain barrier or other barriers within the body. |
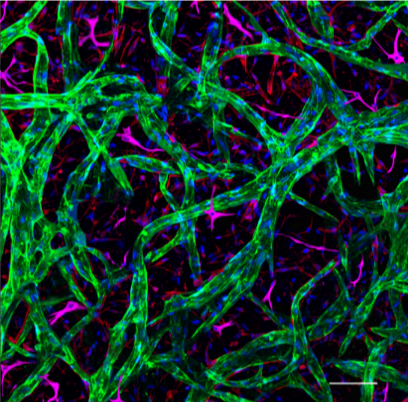
| Blood-brain barrier model. Endothelial networks (green) form by self-assembly into a complex microvascular network with permeabilities comparable to the blood-brain barrier when co-cultured with astrocytes (magenta) and pericytes (not shown). | |
|
References: |
|
|
|
Neurological Disease Models
| Neurological diseases are increasing in prevalence due to our aging population and lack of effective treatments. Microfluidic models of these systems hold considerable promise for simulating the disease process and probing new approaches to treatment. We currently have models for ALS, SMA and Alzheimer’s disease that we are using for mechanistic studies and drug screening. | |
|
|
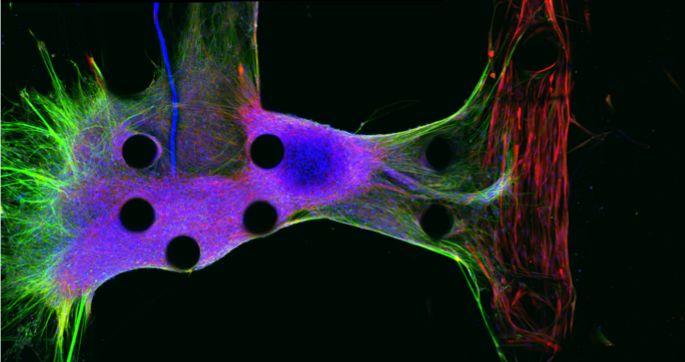
| To create a model of ALS, a neurosphere is generated from human neural precursor cells and seeded into a microfluidic system (left). The neurons then send out neurites (in green) that penetrate into and form neuromuscular junctions inside a muscle strip derived from induced pluripotent stem cells (red). |
|
|
|
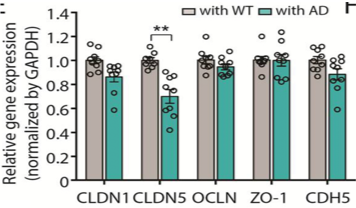
| Human neurons genetically modified to over-express amyloid beta 40 or 42 as a model of Alzheimer’s disease. Here we co-culture the neurons with a vascular barrier to study changes in vessel permeability that occur in cerebral amyloid angiopathy, a vascular condition often associated with and possibly a cause of neuronal death. |
Analysis of gene expression levels of various vascular endothelial cell junctional proteins suggests that the changes in vascular permeability may be caused by a reduction in the expression of claudin-5. |
| References: | |
|
|
Vascularized Organoids (cardiac, cerebral, etc...)
|
Organoid technology, especially using induced pluripoptent stem cells, are the focus of much development for their potential as microphysiological models and in regenerative medicine applications. Currently, however, they lack a perfusable vascular network. This project aims to connect the vascular networks developed in (2) above with networks grown internally in order to enable long-term perfusion. |
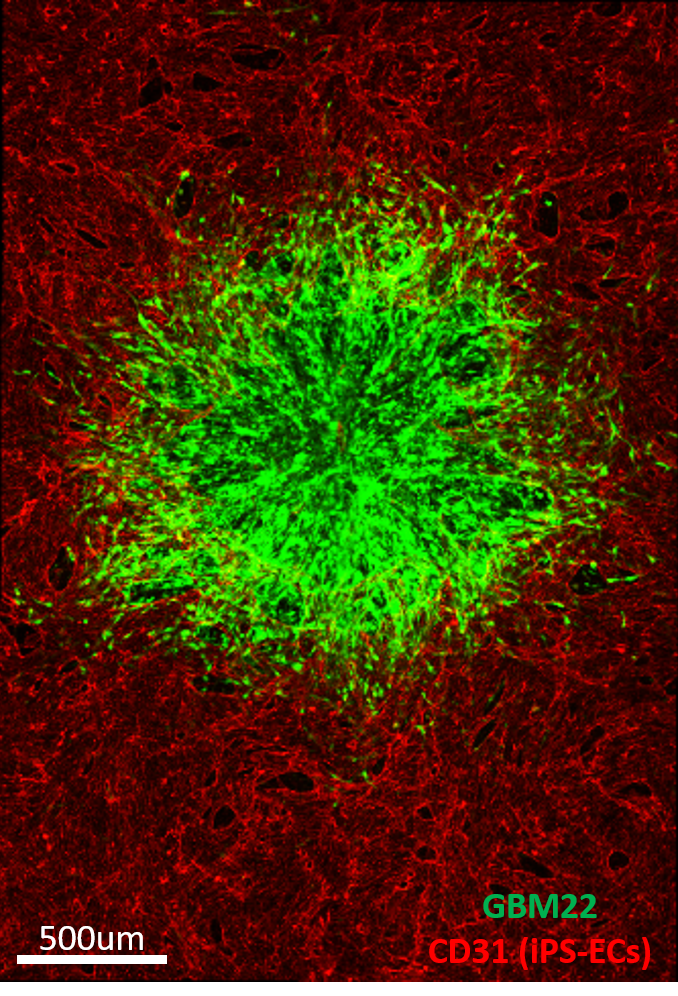
| A tumor (green) is shown immersed in a microvascular network (red) simulating the blood-brain barrier that can be perfused with medium containing drugs or nanoparticles to treat the tumor. This system is used to study the transport of therapeutic agents or immune cells from the vasculature into the tumor. Image by Dr. Cynthia Hajal. | |
|
|
Other Microphysiological Systems and Microfluidic Models
|
As the platform technologies developed in our lab have considerable flexibility, they can be adapted to create models of other organ systems. These include the central and peripheral nervous systems as mentioned above, but also have applications to the reproductive system, the lymphatic circulation, and for cardiac modeling. |
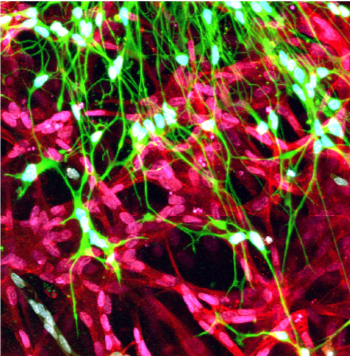
| A co-culture of a cluster of neurons (green) and vascular endothelial cells (red) show interpenetration of the neurites and the vascular network as seen in the brain. | |
|
References: |
|
|
|

