NEURON is an extensible nerve modelling and simulation program. It allows you to create complex nerve models by connecting multiple one-dimensional sections together to form arbitrary neuron morphologies, and allows you to insert multiple membrane properties into these sections (including channels, synapses, and ionic concentrations). The interface was designed to present the neural modeller with an intuitive environment and hide the details of the numerical methods used in the simulation.
This tutorial is divided into 5 parts (A - E) and will take you, step by step, through the process of creating a complex simulation. In part A we start with the basics: how to create a single compartment neuron model with Hodgkin-Huxley conductances, how to run the simulator and how to display the simulation results. In part B we move into the more advanced topics of building multi-compartmental neurons and using different types of graphs to display the results. In part C we will replicate neurons using templates and connect these neurons together. In part D we will add new membrane mechanisms to the simulator and incorporate them in our neurons. Finally, in part E we will look at ways of saving data from the simulations and methods for increasing simulation speed.
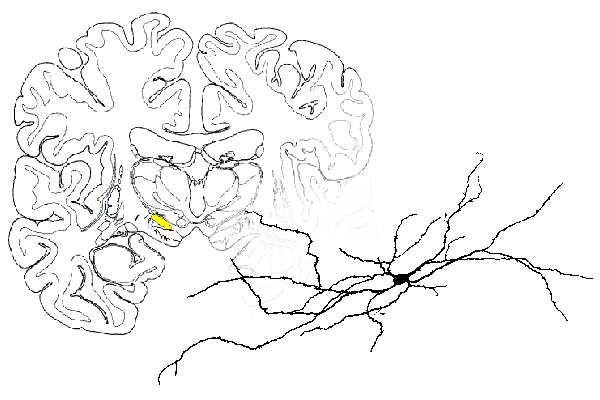
It is always good to have a final product in mind when we start the modelling task, so here is ours:

The equivalent human nucleus is drawn above in coronal section.
All of these neurons have basically the same morphology: a cell body with dendritic trunks with complex branching. We believe there are Hodgkin-Huxley type ion channels in the soma, but are uncertain what type of channels are in the dendrites.
Commands are written in typewriter font. When the command should be entered at the system (UNIX or LINUX) prompt it is in brown type face. When it should be entered at the NEURON prompt (or as part of a hoc program; see later) it is in green type face. The dark blue type face in part D of the tutorial indicates the NMODL language.
When you need to do something (as opposed to just read), the text will be set against a highlighted background. E.g.:
type this
means "type this at the command line or add it to your program".
How you start NEURON depends on what operating system you're using.
nrnguiat the operating system command line prompt.
If everything is installed properly, you should see a "welcome" message that states the version and release date of NEURON, some additional comments, and then a prompt that looks like:
oc>
(Under MSWin and MacOS, a new window pops up that contains these items.) A new window also appears entitled "Main Menu" that provides access to the graphical user interface (outlined below).
When you see the oc> prompt, you can enter NEURON commands directly from the keyboard.
To exit from NEURON, either type quit() at the oc> prompt, or use File/Exit from the interpreter window's drag bar (MSWin and MacOS). An alternative that works only under UNIX is to type CTRL-D at the oc> prompt.
All of the commands in these tutorials can be entered directly from the keyboard, but this would entail quite a bit of typing. A more practical approach is to enter the commands into a program using your favourite editor (e.g. xemacs, notepad) and save them to a plain text file.
This program can then be run when NEURON starts. For example, imagine you have a program called sthA.hoc that you want to run. How to do this depends on your operating system:
nrngui sthA.hocat the system prompt and NEURON will launch and read your program file.
We suggest that you create a file called sthA.hoc and type the commands highlighted in grey into it as you go along. To test what you've done you can always restart NEURON as above.
The first step in creating our model is to build the neuron's morphology. Neurons can usually be described by reducing their complex structures into unbranched cylindrical sections. The simplest reduction is to remove the neuron's axon and dendrites leaving only the cell body. By modelling only the cell body, you possibly lose a tremendous amount of useful information, but you can always increase the morphological complexity as needed (which we will do in parts B and C).
From experimental measurements, we know that our soma is approximately spherical with a diameter of 18.8µm. In NEURON, the neuron's geometry is described in terms of cylindrical sections. Before we set the section's properties, we must first create a new section, soma, as follows:
create soma
This command creates a new section with the default properties (the number of segments in the section [1], the diameter [500 µm], the length [100 µm], the specific membrane capacitance [1 µF/cm2] and the cytoplasmic resistivity [35.4 ohm cm]). For our simulation, we need to change some of our soma's properties; the diameter of the section (diam), the length of the section (L), and the axial resistance (Ra), which in the rat subthalamic nucleus has a high value of 123 ohm-cm. All of these values are taken from data specific to subthalamic projection neurons.
The parameter nseg specifies the number of internal points at which NEURON computes solutions, i.e. the time course of membrane current, potential, ionic concentrations etc.. We can imagine that a section is broken into nseg compartments of equal length (L/nseg), and that NEURON will compute the time course of these variables at the centre of each compartment. These compartments are called segments. If we assume that the density of transmembrane current is reasonably uniform over the surface of the soma, a single point will be sufficient, and we can assign a value of 1 to nseg.
Since NEURON deals with many different sections each with their own unique name, we must tell NEURON which section we are working on when we want to refer to the value of a section parameter. There are three ways to do this in NEURON.
Use the dot notation. Here, we refer to the section properties with the section name followed by a "dot" or period (".") followed by the property name. To assign values to section parameters such we can use:
soma.nseg = 1 soma.diam = 18.8 soma.L = 18.8 soma.Ra = 123.0
To print the value of any section parameter we can use the print command with the dot notation, for example:
print soma.diam
Place the name of the section on the same line and immediately in front of a statement that refers to a section property:
soma nseg = 1 soma diam = 18.8 soma L = 18.8 soma Ra = 123.0
We can group multiple properties within braces:
soma {
nseg = 1
diam = 18.8
L = 18.8
Ra = 123.0
}
The statements within this pair of braces pertain to soma.
Use an access statement to declare a "default" section:
access soma
The access statement tells NEURON that when we aren't specifying a particular section (i.e. we're not using method 1 or 2), all subsequent references to properties of a section belong to the default section, in this case the soma. For example
nseg = 1 diam = 18.8 L = 18.8 Ra = 123.0 print nseg
would change and print the properties of the default section, in this case the soma.
Method 3 is a convenient way of changing properties from the hoc interpreter, but it is generally not the best way of changing section properties in hoc files. This is because it tends to make them harder to read since it is not immediately clear which section a property refers to. However we do need one (but only one) access statement in a model specification:
access soma
We need the access statement because many of NEURON's graphical tools don't work unless a default section has already been declared. We shouldn't have more than one access statement (either in a program or at the oc> prompt) because it's easy to get confused about what section we're referring to. For example, we might type in print v, then type access dend, then (a few dozen commands later) type print v again, forget that we'd changed the default section to dend and think that we were looking at soma. (This problem is worse with the graphical user interface we'll meet later.) We can specify any section we like as the default section, but in general it should be a section that is of special interest.
For part A we mainly will use the second method. Thus, the program so far is:
create soma access soma soma nseg = 1 soma diam = 18.8 soma L = 18.8 soma Ra = 123.0
One point to notice about the program is that the access statement is straight after the create statement. When writing hoc code it's a good practice to group all the create statements together, and put the access statement immediately after the last create statement. This makes sure that there's only one access statement in the program.
Each section in NEURON has the default properties (see above) automatically inserted, but other mechanisms (e.g., channels) with their own properties must be explicitly inserted into a section. NEURON includes two built-in channel membrane mechanisms: Hodgkin-Huxley channels (hh) and passive channels (pas). Each of these mechanisms can be inserted using the insert command. If you have set a default section, you can insert a new mechanism it with:
insert hh
You can also use the method 2 above to insert a new mechanism. E.g. to insert a passive channel in the soma you would type
soma insert pas
You can add as many properties into each section as you need for your model.
When you add a new membrane mechanism to a section, you add the new membrane mechanism's properties and their default values to the section. For example, if you add passive channels to the section, you will introduce two new properties to the section: g_pas (specific membrane conductance [S/cm2]) and e_pas (reversal potential [mV]). For our simulation, neither the default Hodgkin-Huxley channels nor the passive properties will accurately model our neuron type. However, for the moment we will insert these properties to explore a working system, then in part D of the tutorials we will develop our own active channel properties using the model description language (NMODL).
The Hodgkin-Huxley channels add the following new properties to the section:
It also adds the following state variables, that can be displayed with the print command:
To display all of the properties and their current values in a particular section, you can call the psection function. To see the properties of the default section:
psection()
or for a particular section:
soma psection()
or to list the properties of all sections in the model:
forall psection()
For our model, we can use the default values for the other properties. Thus, our program looks like:
create soma access soma soma nseg = 1 soma diam = 18.8 soma L = 18.8 soma Ra = 123.0 soma insert hh
NEURON makes the distinction between mechanisms that are attributed to an entire section (e.g., HH channels) and mechanisms that are associated with a particular point in the section (e.g., voltage clamp or synapse). While the former are most conveniently expressed in terms of per unit area, the point processes are more conveniently expressed in absolute terms (e.g., current injection is usually expressed in terms of nA instead of nA/cm2). Point processes also differ in that you can insert several in the same segment.
In NEURON, point processes are handled as objects which means that to create one you need to first create an object variable to be associated with the object and then create a new object. To declare object variables, you enter the following:
objectvar stim
This creates an object variable named stim, and now you need to create the actual object. Newly created objects need to be associated with a particular section, so we need either to have a default section or to specify the section name with which the object is to be associated. The optional section name is give first followed by the assignment of the object variable to a new instance of a particular object (in this case a current clamp) with the location of the object in the section given in parentheses. The location is specified with a number between 0 and 1 (inclusive) where the number represents the fractional length along the section to place the point process. For example:
stim = new IClamp(0.5)
or with the section name:
soma stim = new IClamp(0.5)
Since the default section in our program is soma, both of these commands will accomplish the same result (i.e., creating a new current clamp object in the middle of the soma).
There are several built-in point processes, including: IClamp, VClamp and ExpSyn. Additional point processes can be built into the simulator with the model description language. As with channels, each point process has its own set of properties. Below is a list of IClamp point processes properties.
IClamp:
In our model, we want to stimulate the soma by giving it a current pulse. We can accomplish this by adding a IClamp as above and then setting its properties. Point processes (and other objects) differ from sections in that there is no concept of a default point process that we can choose using the access statement. Thus, we cannot set the properties in all the same ways we set them in sections. Rather, we must use the dot notation:
stim.del = 100 stim.dur = 100 stim.amp = 0.1
This creates an electrode current clamp in the centre of the soma that will start injecting a 0.1nA current at 100ms for a duration of 100ms.
So, with the IClamp now our program looks like:
create soma access soma soma nseg = 1 soma diam = 18.8 soma L = 18.8 soma Ra = 123.0 soma insert hh objectvar stim stim = new IClamp(0.5) stim.del = 100 stim.dur = 100 stim.amp = 0.1
To ease the process of running simulations, NEURON has a standard run library of useful functions. These library functions are used to initialise the simulator, begin and end simulations, and plot variables versus time or space during a simulation run. The most useful of these functions is the run() function. It initialises and runs a simulation.
There are many ways to load the standard run library. Probably the most convenient is:
load_file("nrngui.hoc")
because it also loads the library that contains NEURON's GUI tools, which are particularly useful during initial program development. MSWin users who run hoc files which contain this command will notice a window entitled "Main Menu" appear; UNIX, LINUX, and MacOS users will already have this window.
Insert this statement at the beginning of your hoc file, then restart NEURON and have it load your program as explained earlier. At the oc> prompt, enter run() to run the simulation. After the simulation completes, NEURON returns you to the oc> prompt. Our program does not generate any output, so nothing will be printed, but if you want to see how variables change, for example, voltage in the soma, you can print variables with the print command:
print soma.v
or just:
print v
since soma is the default section. This is the value at the end of the simulation time. As the default, simulations run for 5 ms of simulated time, but this can be changed by setting the tstop variable to another value (in ms). For example, to run a simulation for 300ms, enter:
tstop = 300 run()
This will set the simulation stop time to 300ms and run the simulation from the beginning.
As mentioned before, the load_file("nrngui.hoc") ensures that we have a "Neuron Main Menu" window on the screen:
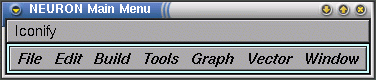
This is the NEURON Main Menu toolbar, which can be used to pop up graphical tools for controlling, displaying, and analysing model parameters and simulation results. The window contains menus File, Edit, Build, Tools, Graph, Vector and Window. For our purposes here we will explore the GUI methods for running a simple simulation. Under the Tools menu select RunControl. This generates a window that allows you to control primary parameters for running the simulation, such as the parameter tstop described above:
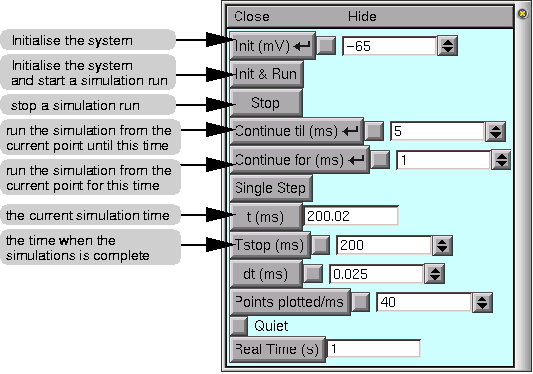
The RunControl window, as with many other windows you will encounter, has a common format. There are buttons, such as "Init & Run" which will run the simulation, and field editors next to some buttons (e.g. next to our tstop variable button). Whenever you see this type of box, you can enter a new value by simply placing the mouse pointer into the field editor, clicking the left mouse button, and typing. A vertical bar should appear in the field editor, indicating the point where what you type will be seen. After you have entered the new number (or expression), you need to tell NEURON to accept the number (or expression) you just entered. You can do this either by pressing the Enter key on your keyboard, or by clicking on any button in the RunControl panel. Some labelled buttons have a smaller, unlabelled square button just to their right. This is a check box, and a red check mark will appear in it if the value in the adjacent field editor has been changed from its default. Thus, if you change the value of tstop, a check mark will appear in the check box signifying that you have changed the value of this field editor. In addition, by pressing the check box, you can toggle between the default value and the changed value.
 In addition to controlling the simulation we will want
to observe the voltage changes in the soma over the period of
simulation (300ms in our current example). Under the
Graph menu of the main window, select Voltage
axis. This generates another window that will display
voltage. When there is more than one section (e.g. when we have
added dendrites - outlined in part B) then we must specify
which section voltage to plot. In the current example the soma
is the default section, so the soma voltage will be plotted by
default. You can open as many voltage or other graph windows as
you like.
In addition to controlling the simulation we will want
to observe the voltage changes in the soma over the period of
simulation (300ms in our current example). Under the
Graph menu of the main window, select Voltage
axis. This generates another window that will display
voltage. When there is more than one section (e.g. when we have
added dendrites - outlined in part B) then we must specify
which section voltage to plot. In the current example the soma
is the default section, so the soma voltage will be plotted by
default. You can open as many voltage or other graph windows as
you like.
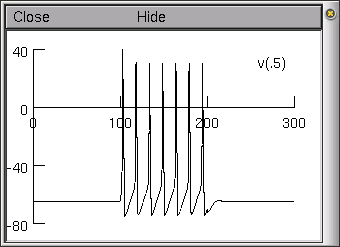
Now, run the simulation (by clicking on the Init & Run button in the RunControl window), observing the voltage being graphed. You should end up with a voltage graph similar to that illustrated here.
So our total program, that builds a subthalamic nucleus soma with an electrode current injection and a graphical user interface, is:
load_file("nrngui.hoc")
create soma
access soma
soma nseg = 1
soma diam = 18.8
soma L = 18.8
soma Ra = 123.0
soma insert hh
objectvar stim
soma stim = new IClamp(0.5)
stim.del = 100
stim.dur = 100
stim.amp = 0.1
tstop = 300
If this was saved as a file, for example sthA.hoc then, as described above, we could start our subthalamic simulation by loading our file into NEURON. For example, on LINUX we use the command:
nrngui sthA.hoc
Next: part B of the tutorial.