|
|
|
Maheshri Research Group
Dynamics and variability in gene regulation and function
Research projects in the lab span a range of topics, including chromatin and transcription dynamics, synthetic biology and genetic regulatory networks, and directed evolution and DNA repair/mutagenesis. Variability is a a common theme as we endeavor:
- to exploit phenotypic variability in gene expression within a population to better understand and engineer promoters and regulatory networks with particular dynamical properties
- to introduce genetic variability in expression and sequence of multiple genes for the directed evolution of multigenic traits
A useful organizational principle is the time scale of the variability -- more specifics of projects organized in this way are detailed below.
Variability and dynamics in gene expression at short time scales
Gene expression tends to be a stochastic, noisy affair. Models of genetic regulatory networks account for this to varying degrees. We are interested in understanding when the stochastic nature of gene expression leads to qualitative differences in predicted behavior. Moreover, how detailed do our stochastic models have to be to essentially "get the answer right"? Does cell growth and division matter and if so, how? Are the dynamics of gene expression at a steady-state really the same as their kinetic response (as is often assumed in models)? How does promoter and chromatin architecture influence these dynamics? To probe these questions, we are developing new experimental tools and computational models as well as utilizing more common experimental techniques.
Current Personnel: C.J. Zopf, Katie Quinn, Nick Wren
Variability and dynamics due to epigenetic gene regulation
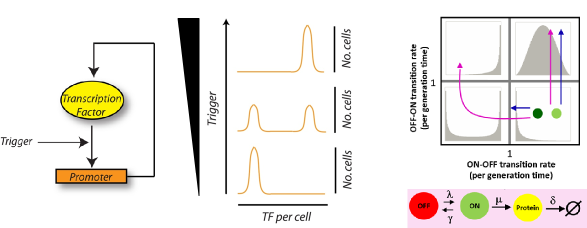
Genes subject to epigenetic regulation switch their expression state on time scales longer than cell division. Such epigentic switches can be broadly classified -- cis- or trans-encoded switches based on the molecular state that encodes the heritable epigenetic mark. We have been studying the dynamics of two epigenetic switches -- a trans-encoded synthetic positive feedback loop and the cis-encoded FLO11 gene. We are particularly interested in understanding the molecular events surrounding a switch event and the how fast fluctuations in these events can lead to slow switching between two macroscopic gene expression states.
People: Katie Quinn, Richard Joh
Variability and dynamics in gene regulation due to genetic variability
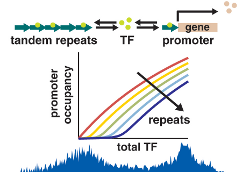
Repetitive regions in genomes can grow and shrink in length on relatively short evolutionary timescales. Along with our collaborators, we are interested in whether these quantitative changes region length can lead to qualitative changes in gene expression and regulatory networks. In fact, our work suggests that if such regions contain transcription factor binding sites, they can serve as decoys, competing with promoters for the transcription factor and altering gene expression. Interestingly, it appears that the residence time of these transcription factors bound to a promoter binding site is significantly shorter than on decoy sites. We are also interested in using genetic variability, particularly in promoter regions, to better understand regulatory networks.
People: Tek-Hyung Lee
Engineered genetic variability for directed evolution of multigenic phenotypes
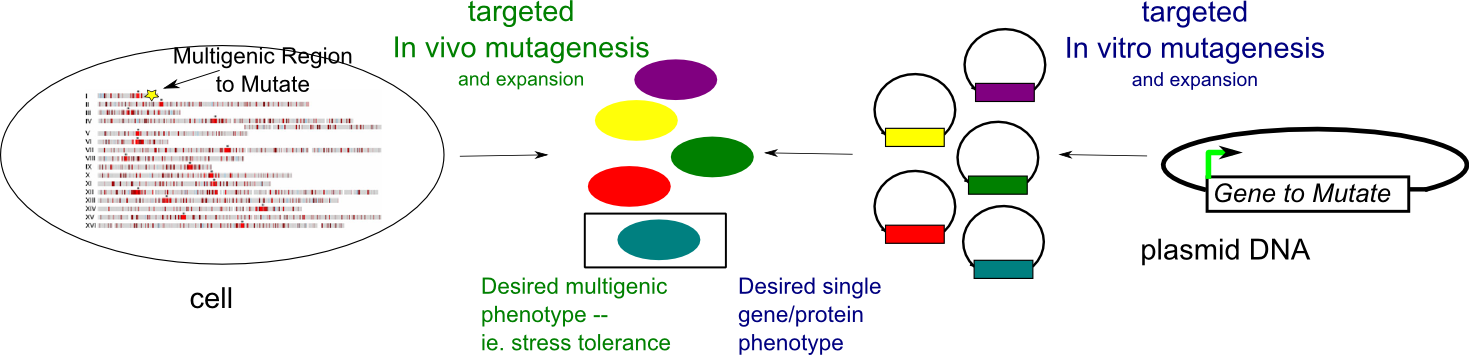
Many industrially-relevant traits of (micro)organisms depend on multiple genes. For example, improved tolerance to environmental stresses in yeast involves many regulatory and structural genes. Directed evolution has been a successful strategy to improve a a protein's properties even in the absence of detailed structural and mechanistic information. Typically, the gene encoding the protein is encoded is mutagenized in vitro, introduced into an organism,and the resulting library is screened/selected for the relevant trait. Unfortunately, this approach is not easily scaleable to many genes and requires the ability to incorporate foreign genetic material into an organism with high efficiency. We have developed a method to target mutagenesis to a specific genomic region spanning 20-30 kb, elevating mutagenesis several orders of magnitude in the region, but not affecting mutation rates elsewhere. We are currently testing whether selectively mutating multiple genes in a pathway can yield desired traits related to that pathway versus classical approachs of mutagenesis in vivo which indiscriminately mutate the genome. Because modulating gene expression rather than gene sequence might be important for multigenic phenotypes, we are also designing genetic strategies to randomly modulate expression levels of multiple genes.
People: Shawn Finney-Manchester, Bradley Niesner