StarGenetics User Manual
Learn how to set up and analyze experiments in
StarGenetics.
Contents
-
Welcome to StarGenetics
-
Opening
StarGenetics
-
StarGenetics at a
glance
-
Fly
exercises
-
Punnett Square
tool
-
Yeast
exercises
-
Renaming
experiments & organisms
-
Matings
involving progeny
-
Discarding experiments & removing organisms
-
Saving
experiments
-
Exporting data
-
Reporting
bugs or asking questions
Welcome to
StarGenetics
StarGenetics provides a set of tools for analyzing
genetic traits. This software simulates mating
experiments between organisms that are genetically
different across a range of traits and allows
students to analyze the nature of the traits in
question. StarGenetics runs on standard Linux,
Windows and Mac computers.
Opening
StarGenetics
To get started with StarGenetics:
- Navigate to http://web.mit.edu/star/genetics.
- Click on the Start button to launch the
StarGenetics application.
- Click Trust
when a prompt appears asking if you trust the
certificate.
- Click on File ->
New in the drop-down menu.
- On the Welcome page choose an
exercise and click on it. To open an exercise that is
not bundled with StarGenetics, click on
File ->
Open and then click on the desired file.
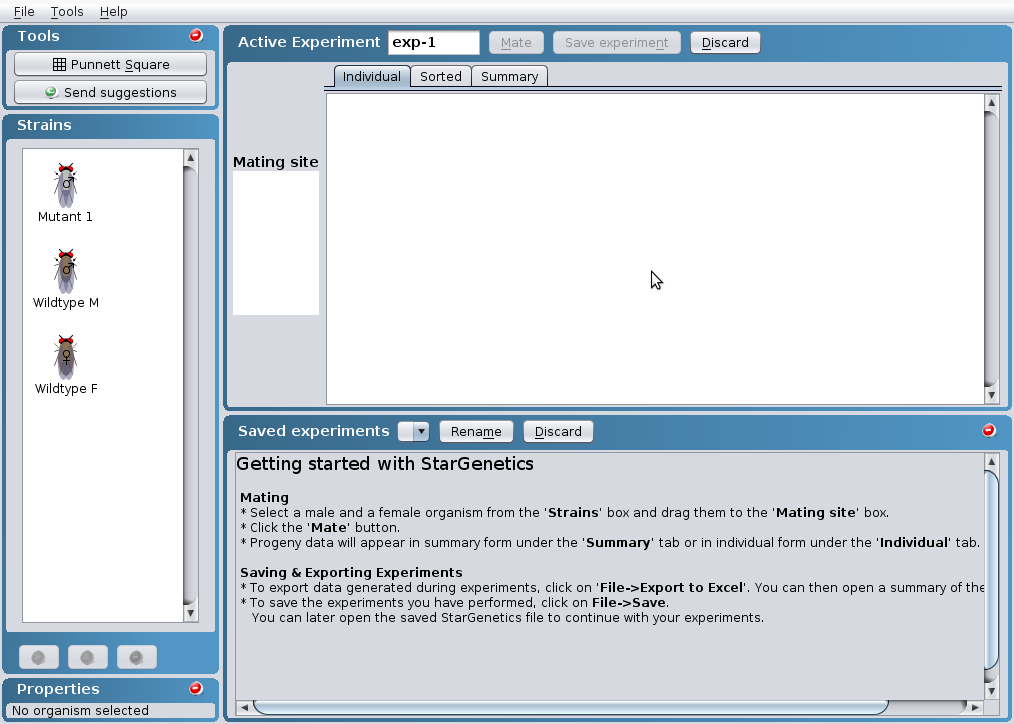
StarGenetics
at a glance
This is the view you will see when you open an
exercise in StarGenetics.
Fly Exercises
Mating Experiment
To perform a mating between two organisms:
- To mate two organisms you can use one of the
following 2 methods:
- drag each organism to the Mating site using the
mouse.
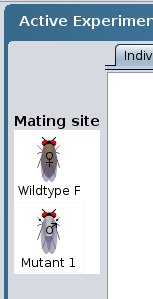
-OR-
- click on the organism and then click on the
Set as a
Parent icon at the bottom of the
Strainsbox.

- Next, click on the
Mate button.
- Mating results can be viewed individually under
the Individual
tab, in sorted form under the Sorted tab, or in summary
form under the Summary tab.
Individual results: progeny listed
individually
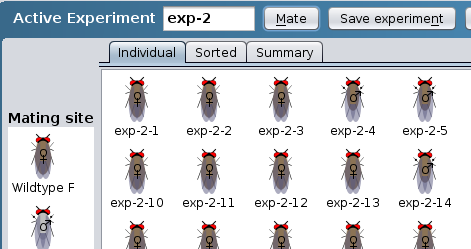
Sorted results: progeny sorted by
sex and phenotype
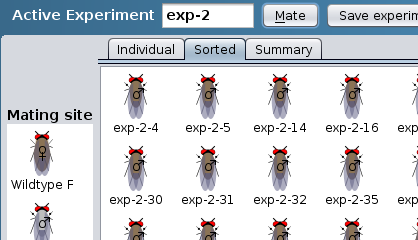
Summary results: progeny listed by
phenotype
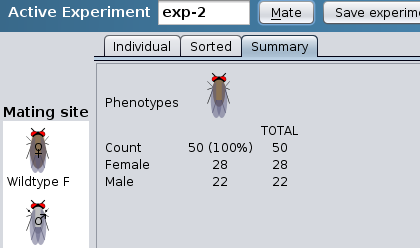
- To start a new experiment click on the
Save experiment
button.
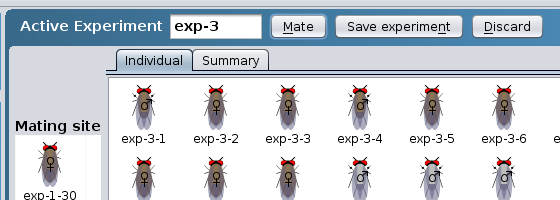
Old experiments will move to the Saved experiments area.
Each tab within the Saved experiments area
represents a prior experiment. Any action that
can be performed within the Active experiment box
can also be performed for a past experiment with
the exception of adding more matings to the
experiment (see bellow in “Tip – carrying out
multiple matings from the same cross”).
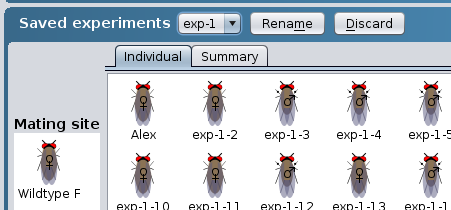
Tip - carrying out multiple matings from
the same cross
To add additional progeny to a mating:
(you might want to do this to increase the
statistical significance of your results)
- Click on the Mate button next to the Mating
site or on the tool bar.
- A prompt will appear asking if you like to add
more progeny to that particular mating. Click Yes.
The number of matings that an organism can perform
can be restricted. When this property is specified,
the number of remaining matings is reflected in the
organism properties section.
Matings
involving progeny
To reserve a progeny for a future
mating:
Reserve an organism from a mating result for a future
experiment by doing one of the following:
- Fly exercises: with the Individual tab selected,
drag the organism to the Strains box.
- Yeast exercises: drag one of the individual
tetrads to the
Strainsbox.
-OR-
- click on the organism/tetrad and then click on
the Add to
strains icon at the bottom of the
Strains
box.

To directly use a progeny for a new
experiment:
You can also use a progeny from a past experiment for
a future experiment without reserving it and adding
it to the Strains box. Click on the
progeny and drag it to the Mating site or set is as
parent as previously described in section “Mating
organisms.”
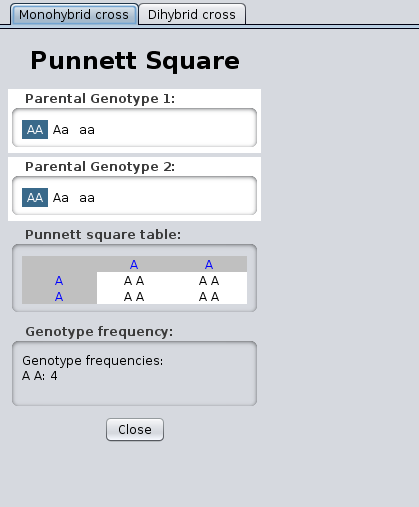
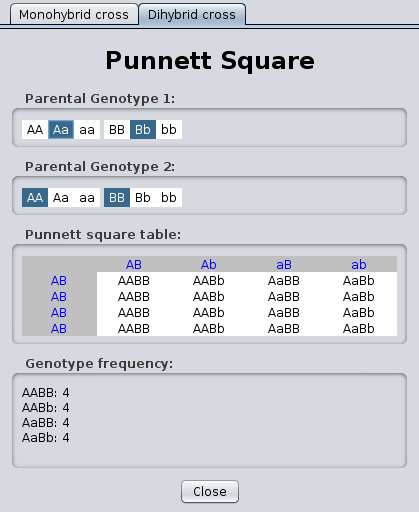
Punnett Square tool
You can use the Punnett
Square tool to help decipher the genotype of a
particular organism after performing a genetic cross.
The Punnett
Square tool allows you to determine the
expected genotypic distribution of the progeny given
the genotypes of the parents.
To use the Punnett Square tool:
- Click on the Punnett Square button in the
Tools box.
- Select a genotype for each parent:
- AA: homozygous dominant
- Aa: heterozygous
- aa: homozygous recessive
- The genotypic frequencies for the cross are
summarized at the bottom of the tool.
- You can view a monohybrid cross or a dihybrid
cross:
Yeast Exercises
When you open a yeast exercise, you have two choices
for experimental set up:
- Non-tetrad experiment
- Tetrad experiment
Non-tetrad experiment
This type of experimental set up allows you to:
- Test individual or multiple strains in selective
media.
- Perform a complementation test with one or more
strains at a time.
Tetrad experiment
This type of experimental set up allows you to:
- Mate two strains at a time and perform a simple
complementation test.
- Mate two strains and sporulate to generate
tetrads for tetrad analysis.
- Mate tetrads to other strains, including MATa or
MATalpha tester strains to deduce tetrads' mating
type.
Experiments
using the Non-tetrad experimental setup
- Drag a strain to the Drop strain here box. The
Drop strain here box moves down to allow the addition
of other strains to the same test.
- Select
- a lawn within the Select lawn box containing
one of the available strains + the appropriate
media within the Select media box to perform a
complementation test
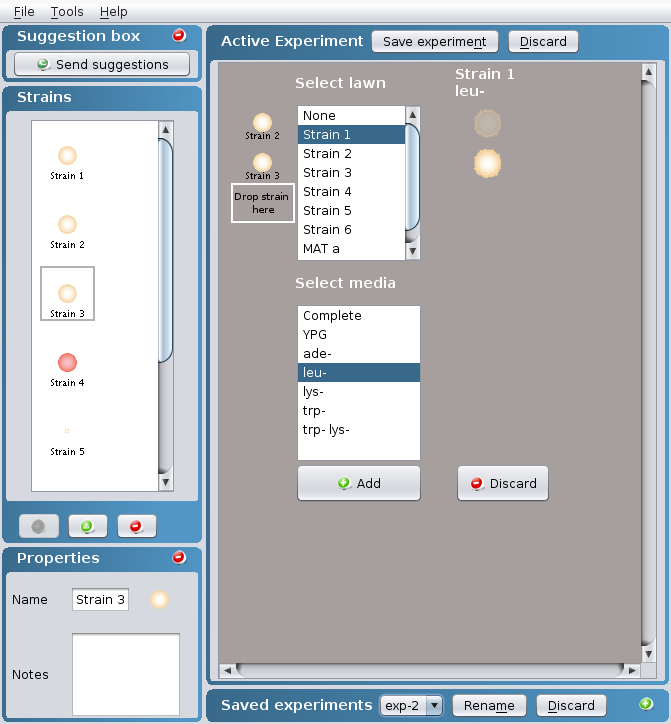
OR
- the appropriate media within the Select media
box in which you would like to replica plate your
strain(s) to test the strain(s) phenotypes. More
than one media can be selected at a time by
clicking on Crontrol + Command (PC) or Control +
Apple (Mac).
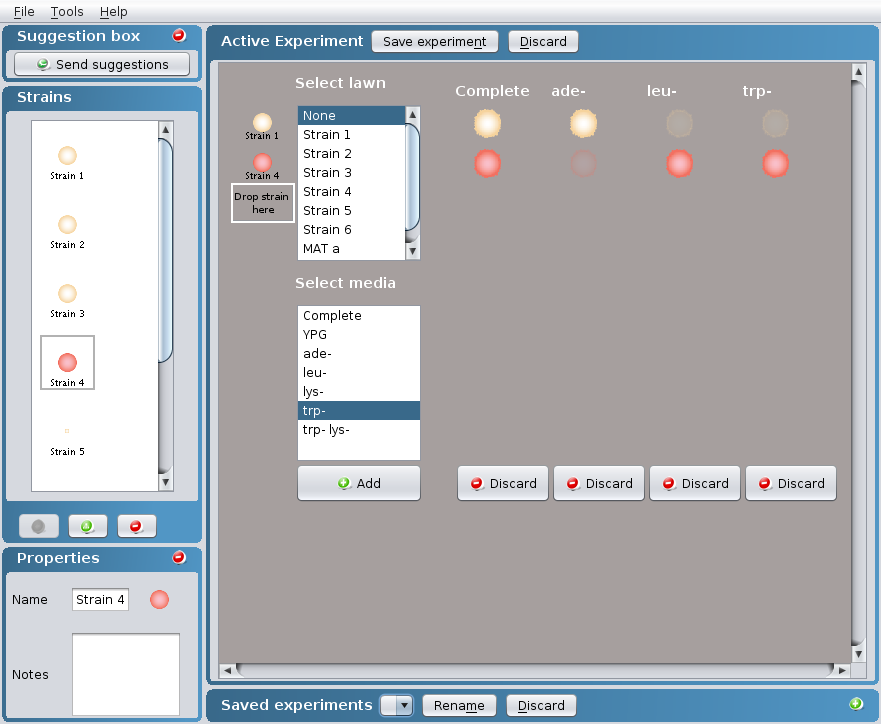
- Click on the Add button at the bottom of the
Select media box.
- Additional experiments can be performed by
selecting the appropriate lawn and/or media and then
clicking on the Add button.
Experiments using
the Tetrad experimental setup
To perform a mating between two
strains
- To mate two strains you can use one of the
following 2 methods:
- drag each strain to the Drop here to mate
box
OR
- click on the strain and then click on the Set as
a Parent icon at the bottom of the Strains box.
- The two strains will automatically mate to
produce a diploid.
- You can either
- replicate plate the newly created diploid to do a
simple complementation test by selecting the
appropriate media within the Select media box.
Click on the Add button at the bottom of the Select
media box. More than one media can be selected at a
time by clicking on Crontrol + Command (PC) or
Control + Apple (Mac).
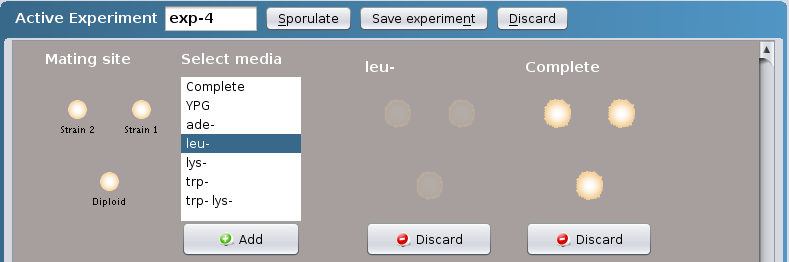
OR
- sporulate the diploid to perform tetrad analysis
by clicking on the Sporulate button.
To conduct tetrad analysis
- Once tetrads are generated (see "To perform a
mating between two strains" section), you can perform
tetrad analysis by selecting the appropriate media
within the Select media box in which you would like
to replica plate your tetrads. More than one media
can be selected at a time by clicking on Crontrol +
Command (PC) or Control + Apple (Mac).
- Click on the Add button at the bottom of the
Select media box.
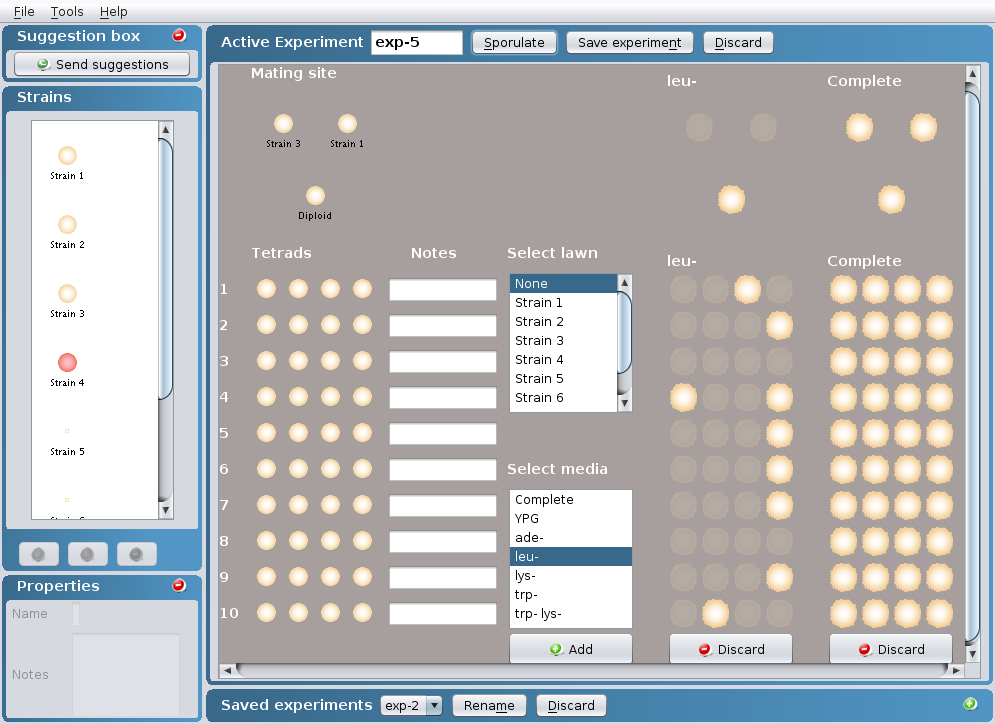
- To mate the generated tetrads to one of the
available strains, select the appropriate strain
within the Select lawn box + the appropriate media
within the Select media box and click on the Add
button at the bottom of the Select media box.
Tip - Annotating Tetrads
To help you keep track of the types of tetrads
generated in your matings as you perform your
tetrad analysis, click on the Notes text box to the
right of the Tetrads section and enter your desired
annotation for each tetrad.
Renaming
experiments & organisms
To rename an experiment:
- To rename an active experiment, click on the text
field containing the experiment's name
-OR-
To rename a saved experiment, click on the
Rename button
- Enter the new name into the prompt and click
OK.
To rename an organism/strain:
- To rename an individual organism/strain first
click on the organism/strain. The organism's/strain's
properties will appear in theProperties box on the left
hand corner.

- Click the "Name" text field.
- Enter the new name into the prompt and click
OK.
Tip – finding more information about an
organism
For fly exercises, the properties dialog describes
phenotypic characteristics (i.e. body color &
wing size) as well as non-phenotypic
characteristics (i.e. number of matings the
organism can perform). You can use the Note box to write any
relevant information about the organism/strain in
question such as its genotype and the type of
mutation exhibited by the organism/strain (i.e.
dominant vs. recessive). Notes entered will appear
to the right of the organism/strain within the
Strains box.
Discarding
experiments & removing organisms
To discard an experiment:
- Click on the Discard button next to the
Mating button
(Fly exercise) orSave
experiment button (Yeast
exercise).
- A prompt will appear asking if you are sure you
want to discard the experiment.
- Click OK.
To remove an organism:
To remove an organism/strain from the Strains box, click on the
organism/strain and then click on the Remove icon at the bottom of
the Strains box.

- You can remove an organism/strain from the
Strains box.
- You can't remove an organism/strain from the
Mating site, but
you can replace it with another organism/strain.
- You can't remove an organism/strain from the
resulting progeny of a mating experiment.
Saving experiments
To save your work so that it can be accessed
at a later time:
- Click on File ->
Save.
- In the “Save As” field box, enter your desired
name for the file.
- To access the experiment at a later time, open
StarGenetics. Click on
File -> Open and click on the previously
saved file.
Exporting data
To export the data generated in your
StarGenetics experiments:
- Click on File ->
Export to Excel.
- In the “Save As” field box, enter your desired
name for the file.
- Open the Excel file. The first tab within the
Excel file corresponds to a summary of all your
"Saved experiments". A summary of the "Discarded
experiments" is available in a second tab.
Note: exporting data is currently only available for
"Fly exercises".
Reporting
bugs or asking questions
To report a bug, request a feature, make a
comment,or ask a question on StarGenetics:
- Click on the Send
suggestionsbutton in the Tools box (Fly exercises)
orSendSuggestion box (Yeast
exercises).

- Fill out the feedback form and click on
Send Report.
















![]()
