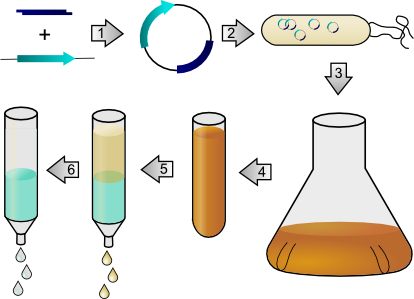
Schemiatic of protein synthesis in a bacterial host.
Proteins will be synthesized using bacterial expression in E. coli host strains. This technique gives us the greatest versatility in preparing protein-based materials because it allows for the individual control of each amino acid in the polypeptide, it allows high molecular weight materials such as whole proteins and protein block copolymers to be synthesized, and with high yield proteins it can be scaled to yield gram quantities of material. In addition, we can use techniques for global amino acid replacement first developed in Prof. Dave Tirrell's Lab to incorporate metal centers (useful for increasing X-ray or electron contrast), fluorinated groups (useful for tuning monomer interaction parameters), or bioorthogonal azide, alkyne, hydrazide, or alkene functionalities (useful for linking proteins and polymers together).
In order to synthesize proteins, a gene encoding for the desired protein is first cloned, and this gene is inserted into an expression vector (step 1). This plasmid is transformed into a strain of E. coli optimized for high-level expression of protein (step 2). The cells are then cultured to produce the desired protein (step 3), and the cells are harvested to collect the unpurified cellular material containing the protein of interest (step 4). The unpurified mixture is mixed with chromatography resins containing immobile metal atoms that bind strongly to a short amino acid tag appended to the end of the synthetic protein (step 5), allowing the impurities to be washed off and pure protein product to be eluted (step 6).