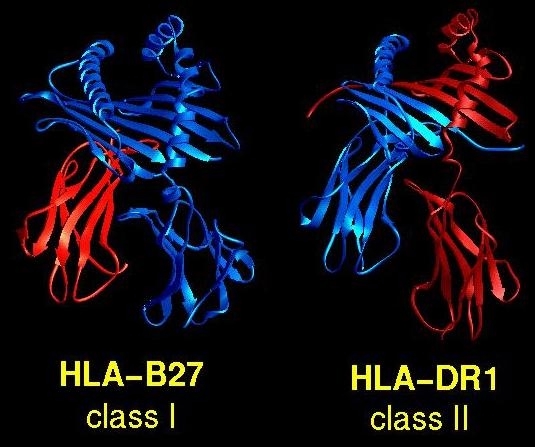
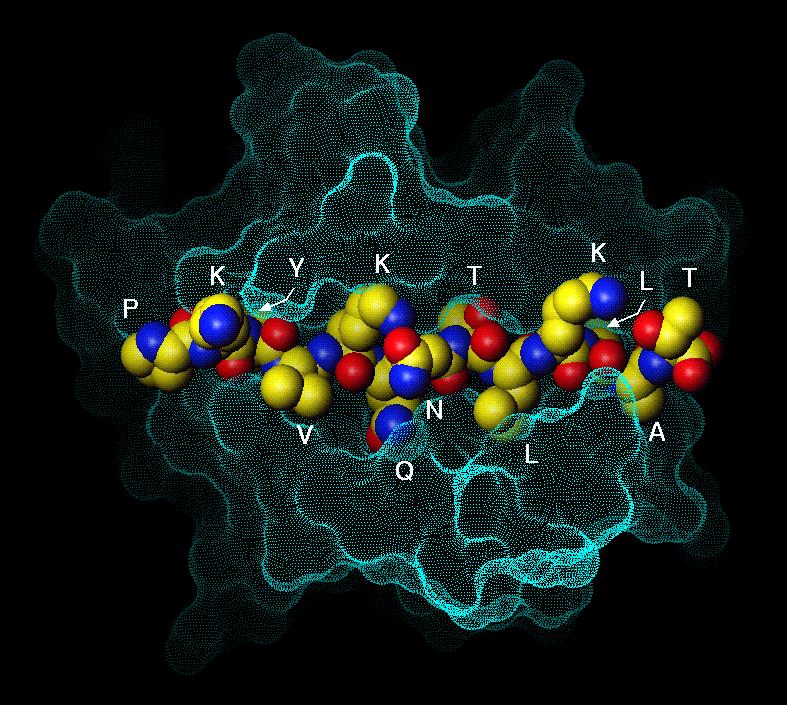
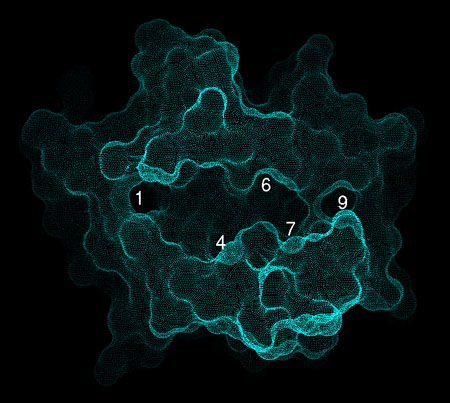
MHC proteins have allele-specific peptide
binding preferences, called motifs
HLA-DR1, our favorite MHC protein, exhibits
side chain preferences at positions 1,4,6,9
position 1: Trp,Tyr,Phe > Met, Leu,
Ile, Val
position 4: Met, Gln, Leu
position 6: Gly,Pro,Ala,Ser
position 9: hydrophobic
Note that these match the positions of
pockets observed in the structure as seen at right
There are several prediction programs available
which give binding scores for different peptide binding registers in a
sequence.
1: EpiPred
by Thomas Cameron
Excel spreadsheets which analyze many
alleles according to algorithms of Hammer/Sinigaglia, A. Sette, and the
Virtual Matrices of Sturniolo and
Hammer, limited to peptides of lenth 1000
aa. **NOTE: this must be run in Internet Explorer.
2: TEPITOPE,
available from J. Hammer at his non-profit company Vaccinome
predicting epitopes according to the Virtual Matrices of Sturniolo and
Hammer.
3: ProPred,
by Harpreet Singh, sponsored by the Bioinformatics
Centre (BIC) at the Institute of Microbial
Technology in Chandigarh, India which predicts epitope peptides based on
the Virtual matrices of Struniolo and Hammer. This has a nice graphical
format for its output.
4: SYFPEITHI
uses matrices which are described in a recent book by HG Rammensee et al.
5: Epipredict
uses matrices developed primarily by B. Fleckenstein and K.-H Wiesmüller.
Try several to see which you prefer.