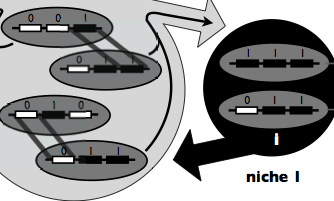
Symsim is a discrete generations model implemented in MatLab, with varying
population size, designed to investigate the colonization of a new
ecological niche. Each generation consists of resource competition,
population growth and recombination in a sympatric pool. Symsim models a
microbial population growing in an environment composed of two distinct
niches, one ancestral (niche 0) and one derived (niche 1). Genotypes consist
of L unlinked adaptive loci, each with two allelic states, 0 or 1,
conferring adaptation to niche 0 or 1, respectively.
Download code and documentation
The symsim model is described in detail in the paper:
Friedman J, Alm EJ, Shapiro BJ. (2012) Sympatric speciation: when is it
possible in bacteria? PLOS ONE. (accepted Dec. 2012)
Summary of the paper:
It has been theorized for some time that sexual animal populations diverge more
readily into new species when a small number of genes are sufficient to initiate
and complete the speciation process. Some of these genes may promote sexual
isolation between species (important in 'sympatric' speciation, where there are
no physical barriers to mating) and others may confer adaptation to different
ecological niches. Bacteria, like sexual organisms, adapt to diverse ecological
niches. Yet they reproduce clonally, exchanging genes occasionally by
recombination, potentially with distant relatives, such that sexual isolation
between species is never complete. Despite these differences from sexual
organisms, we show that bacteria are also more likely to form new species when
just a few genes are involved in niche adaptation. The reason for this is an
evolutionary tradeoff: frequent genetic exchange (recombination) is required to
generate adaptive combinations of multiple adaptive alleles, but without
mechanisms for sexual isolation these adapted species tend to merge back
together rather than diverging into separate species. This tradeoff imposes a
simple constraint on the speciation process in bacteria, and may explain certain
aspects of bacterial biology such as the clustering of genes into operons
(effectively reducing the number of genes involved in speciation) and variable
recombination rates over time.

 Symsim is a discrete generations model implemented in MatLab, with varying
population size, designed to investigate the colonization of a new
ecological niche. Each generation consists of resource competition,
population growth and recombination in a sympatric pool. Symsim models a
microbial population growing in an environment composed of two distinct
niches, one ancestral (niche 0) and one derived (niche 1). Genotypes consist
of L unlinked adaptive loci, each with two allelic states, 0 or 1,
conferring adaptation to niche 0 or 1, respectively.
Symsim is a discrete generations model implemented in MatLab, with varying
population size, designed to investigate the colonization of a new
ecological niche. Each generation consists of resource competition,
population growth and recombination in a sympatric pool. Symsim models a
microbial population growing in an environment composed of two distinct
niches, one ancestral (niche 0) and one derived (niche 1). Genotypes consist
of L unlinked adaptive loci, each with two allelic states, 0 or 1,
conferring adaptation to niche 0 or 1, respectively.