Thermodynamic Properties
[Models of Phases of Matter]
These classes are used to compute the thermodynamic properties of phases of matter. More...
|
Classes | |
| class | ConstDensityThermo |
| Overloads the virtual methods of class ThermoPhase to implement the incompressible equation of state. More... | |
| class | DebyeHuckel |
| Class DebyeHuckel represents a dilute liquid electrolyte phase which obeys the Debye Huckel formulation for nonideality. More... | |
| class | EdgePhase |
| A thermodynamic Phase representing a one dimensional edge between two surfaces. More... | |
| class | HMWSoln |
| Class HMWSoln represents a dilute or concentrated liquid electrolyte phase which obeys the Pitzer formulation for nonideality. More... | |
| class | IdealGasPhase |
| Class IdealGasPhase represents low-density gases that obey the ideal gas equation of state. More... | |
| class | IdealMolalSoln |
| This phase is based upon the mixing-rule assumption that all molality-based activity coefficients are equal to one. More... | |
| class | IdealSolidSolnPhase |
| Class IdealSolidSolnPhase represents a condensed phase ideal solution compound. More... | |
| class | IdealSolnGasVPSS |
| This class can handle either an ideal solution or an ideal gas approximation of a phase. More... | |
| class | LatticePhase |
| A simple thermoydnamics model for a bulk phase, assuming a lattice of solid atoms. More... | |
| class | MetalSHEelectrons |
| Class MetalSHEelectrons represents electrons within a metal, adjacent to an aqueous electrolyte, that are consistent with the SHE reference electrode. More... | |
| class | MineralEQ3 |
| Class MineralEQ3 represents a stoichiometric (fixed composition) incompressible substance based on EQ3's parameterization. More... | |
| class | MolalityVPSSTP |
| class | PureFluidPhase |
| This phase object consists of a single component that can be a gas, a liquid, a mixed gas-liquid fluid, or a fluid beyond its critical point. More... | |
| class | SingleSpeciesTP |
| The SingleSpeciesTP class is a filter class for ThermoPhase. More... | |
| class | UnknownSpeciesThermoModel |
| Throw a named error for an unknown or missing species thermo model. More... | |
| class | SpeciesThermoFactory |
| Factory to build instances of classes that manage the standard-state thermodynamic properties of a set of species. More... | |
| class | StoichSubstanceSSTP |
| Class StoichSubstanceSSTP represents a stoichiometric (fixed composition) incompressible substance. More... | |
| class | SurfPhase |
| A simple thermoydnamics model for a surface phase, assuming an ideal solution model. More... | |
| class | UnknownThermoPhaseModel |
| Specific error to be thrown if the type of Thermo mananger is unrecognized. More... | |
| class | ThermoFactory |
| Factory class for thermodynamic property managers. More... | |
| class | ThermoPhase |
| Base class for a phase with thermodynamic properties. More... | |
| class | VPStandardStateTP |
| This is a filter class for ThermoPhase that implements some prepatory steps for efficiently handling a variable pressure standard state for species. More... | |
| class | WaterPropsIAPWS |
| Class for calculating the equation of state of water. More... | |
| class | WaterSSTP |
| Class for single-component water. More... | |
Modules | |
| Species Standard-State Thermodynamic Properties | |
In this module we describe Cantera's treatment of pressure dependent standard states (PDSS) objects. | |
| Managers for Calculating Reference-State Thermodynamics | |
The ThermoPhase object relies on a set of manager classes to calculate the thermodynamic properties of the reference state for all of the species in the phase. | |
| Species Reference-State Thermodynamic Properties | |
The ThermoPhase object relies on classes to calculate the thermodynamic properties of the reference state for all of the species in the phase. | |
| Managers for Calculating Standard-State Thermodynamics | |
To compute the thermodynamic properties of multicomponent solutions, it is necessary to know something about the thermodynamic properties of the individual species present in the solution. | |
Functions | |
| ThermoPhase * | newThermoPhase (std::string model, ThermoFactory *f=0) |
| Create a new thermo manager instance. | |
| std::string | eosTypeString (int ieos, int length=100) |
| Translate the eosType id into a string. | |
| ThermoPhase * | newPhase (std::string infile, std::string id) |
| Create and Initialize a ThermoPhase object from an XML input file. | |
| bool | importPhase (XML_Node &phase, ThermoPhase *th, SpeciesThermoFactory *spfactory=0) |
| Import a phase information into an empty thermophase object. | |
| bool | installSpecies (int k, const XML_Node &s, thermo_t &p, SpeciesThermo *spthermo_ptr, int rule, XML_Node *phaseNode_ptr=0, VPSSMgr *vpss_ptr=0, SpeciesThermoFactory *factory=0) |
| Install a species into a ThermoPhase object, which defines the phase thermodynamics and speciation. | |
| const XML_Node * | speciesXML_Node (std::string kname, const XML_Node *phaseSpeciesData) |
| Search an XML tree for species data. | |
Detailed Description
These classes are used to compute the thermodynamic properties of phases of matter.
The main base class for describing thermodynamic properties of phases within Cantera is called ThermoPhase. ThermoPhase is a large class that describes the interface within Cantera to Thermodynamic functions for a phase.
The calculation of thermodynamic functions within ThermoPhase is broken down roughly into two or more steps. First, the standard state properties of all of the species are calculated at the current temperature and at either the current pressure or at a reference pressure. If the calculation is carried out at a reference pressure instead of at the current pressure the calculation is called a "reference state properties" calculation, just to make the distinction (even though it may be considered to be a fixed-pressure standard-state calculation). The next step is to adjust the reference state calculation to the current pressure. The thermodynamic functions then are considered to be at the standard state of each species. Lastly the mixing contributions are added to arrive at the thermodynamic functions for the solution.
The ThermoPhase class provides interfaces to thermodynamic properties calculated for the reference state of each species, the standard state values for each species, the thermodynamic functions for solution values, both on a per mole of solution basis (i.e., enthalpy_mole()), on a per kg of solution basis, and on a partial molar basis for each species (i.e., getPartialMolarEnthalpies(double *hbar)). At each level, functions for the enthalpy, entropy, Gibbs free energy, internal energy, and volume are provided. So, 5 levels (reference state, standard state, partial molar, per mole of solution, and per mass of solution) and 5 functions multiplied together makes 25 possible functions. That's why ThermoPhase is such a large class.
Categorizing the Different ThermoPhase Objects
ThermoPhase objects may be catelogged into four general bins.
The first type are those whose underlying species have a reference state associated with them. The reference state describes the thermodynamic functions for a species at a single reference pressure, 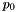 . The thermodynamic functions are specified via derived objects of the SpeciesThermoInterpType object class, and usually consist of polynomials in temperature such as the NASA polynomial or the SHOMATE polynomial. Calculators for these reference states, which manage the calculation for all of the species in a phase, are all derived from the virtual base class SimpleThermo. Calculators are needed because the actual calculation of the reference state thermodynamics has been shown to be relatively expensive. A great deal of work has gone into devising efficient schemes for calculating the thermodynamic polynomials of a set of species in a phase, in particular gas species in ideal gas phases whose reference state thermodynamics is specified by NASA polynomials.
. The thermodynamic functions are specified via derived objects of the SpeciesThermoInterpType object class, and usually consist of polynomials in temperature such as the NASA polynomial or the SHOMATE polynomial. Calculators for these reference states, which manage the calculation for all of the species in a phase, are all derived from the virtual base class SimpleThermo. Calculators are needed because the actual calculation of the reference state thermodynamics has been shown to be relatively expensive. A great deal of work has gone into devising efficient schemes for calculating the thermodynamic polynomials of a set of species in a phase, in particular gas species in ideal gas phases whose reference state thermodynamics is specified by NASA polynomials.
The reference state thermodynamics combined with the mixing rules and an assumption about the pressure dependence yields the thermodynamic functions for the phase. Expressions involving the specification of the fugacities of species would fall into this category of ThermoPhase objects. Note, however, that at this time, we do not have any nontrivial examples of these types of phases. In general, the independent variables that completely describe the state of the system for this class are temperature, the phase density, and 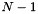 species mole or mass fractions. Additionally, if the phase involves charged species, the phase electric potential is an added independent variable. Examples of the first class of ThermoPhase functions, which includes the IdealGasPhase object, the most commonly used object with Cantera, are given below.
species mole or mass fractions. Additionally, if the phase involves charged species, the phase electric potential is an added independent variable. Examples of the first class of ThermoPhase functions, which includes the IdealGasPhase object, the most commonly used object with Cantera, are given below.
- IdealGasPhase in IdealGasPhase.h
- StoichSubstance in StoichSubstance.h
- SurfPhase in SurfPhase.h
- EdgePhase in EdgePhase.h
- LatticePhase in LatticePhase.h
- LatticeSolidPhase in LatticeSolidPhase.h
- ConstDensityThermo in ConstDensityThermo.h
- PureFluidPhase in PureFluidPhase.h
- IdealSolidSolnPhase in IdealSolidSolnPhase.h
- VPStandardStateTP in VPStandardStateTP.h
The second class of objects are actually all derivatives of the VPStandardState class listed above. These classes assume that there exists a standard state for each species in the phase, where the Thermodynamic functions are specified as a function of temperature and pressure. Standard state objects for each species are all derived from the PDSS virtual base class. Calculators for these standard state, which coordinate the calculation for all of the species in a phase, are all derived from the virtual base class VPSSMgr. In turn, these standard states may employ reference state calculation to aid in their calculations. And the VPSSMgr calculators may also employ SimpleThermo calculators to help in calculating the properties for all of the species in a phase. However, there are some PDSS objects which do not employ reference state calculations. An example of this is real equation of state for liquid water used within the calculation of brine thermodynamcis. In general, the independent variables that completely describe the state of the system for this class are temperature, the phase pressure, and N - 1 species mole or mass fractions or molalities. The standard state thermodynamics combined with the mixing rules yields the thermodynamic functions for the phase. Mixing rules are given in terms of specifying the molar-base activity coefficients or activities. Lists of phases which belong to this group are given below
Note, the ideal gas and ideal solution approximations are lumped together in the class IdealSolnGasVPSS, because at this level they look alike having the same mixing rules with respect to the specification of the excess thermodynamic properties.
The third class of objects are actually all derivatives of the MolalityVPSSTP object. They assume that the standard states are temperature and pressure dependent. But, they also assume that the standard states are molality-based. In other words they assume that the standard state of the solute species are in a pseudo state of 1 molality but at infinite dilution. A solvent must be specified in these calculations. The solvent is assumed to be species zero, and its standard state is the pure solvent state. Lists of phases which belong to this group are:
The fourth class of ThermoPhase objects are stoichiometric phases. Stoichiometric phases are phases which consist of one and only one species. The class SingleSpeciesTP is the base class for these substances. Within the class, the general ThermoPhase interface is dumbed down so that phases consisting of one species may be succinctly described. These phases may have PDSS classes or SimpleThermo calculators associated with them. In general, the independent variables that completely describe the state of the system for this class are temperature and either the phase density or the phase pressure. Lists of classes in this group are given below.
The reader may note that there are duplications in functionality in the above lists. This is true. And, it's used for the internal verification of capabilities within Cantera's unit tests.
Setting the State of the phase
Typically, the way the ThermoPhase object works is that there are a set of functions that set the state of the phase via setting the internal independent variables. Then, there are another set of functions that query the thermodynamic functions evalulated at the current State of the phase. Internally, most of the intermediate work generally occurs at the point where the internal state of the system is set and not at the time when individual thermodynamic functions are queried (though the actual breakdown in work is dependent on the individual derived ThermoPhase object). Therefore, for efficiency, the user should lump together queries of thermodynamic functions after setting the state. Moreover, in setting the state, if the density is the independent variable, the following order should be used:
- Set the temperature
- Set the mole or mass fractions or set the molalities
- set the pressure.
For classes which inherit from VPStandardStateTP, the above order may be used, or the following order may be used. It's not important.
- Set the temperature
- Set the pressure
- Set the mole or mass fractions or set the molalities
The following functions are used to set the state:
| setState_TPX() | Sets the temperature, mole fractions and then the pressure of the phase. |
| setState_TPY() | Set the temperature, mass fractions and then the pressure of the phase. |
| setState_TPM() | Set the temperature, solute molalities, and then the pressure of the phase. Only from ThermoPhase objects which inherit from MolalityVPSSTP |
| setState_TP() | Set the temperature, and then the pressure of the phase. The mole fractions are assumed fixed. |
| setState_PX() | Set the mole fractions and then the pressure of the phase. The temperature is assumed fixed. |
| setState_PY() | Set the mass fractions and then the pressure of the phase. The temperature is assumed fixed. |
| setState_HP() | Set the total specific enthalpy and the pressure of the phase using an iterative process. The mole fractions are assumed fixed |
| setState_UV() | Set the total specific internal energy and the pressure of the phase using an iterative process. The mole fractions are assumed fixed. |
| setState_SP() | Set the total specific internal energy and the pressure of the phase using an iterative process. The mole fractions are assumed fixed. |
| setState_SV() | Set the total specific entropy and the total specific molar volume of the phase using an iterative process. The mole fractions are assumed fixed. |
| setConcentrations() | Set the concentrations of all the species in the phase. Note this implicitly specifies the pressure and density of the phase. The temperature is assumed fixed. |
| setDensity() | Set the total density of the phase. The temperature and mole fractions are assumed fixed. Note this implicity sets the pressure of the phase. |
| setTemperature() | Set the temperature of the phase. The density and the mole fractions of the phase are fixed. |
| setToEquilState() | Sets the mole fractions of the phase to their equilibrium values assuming fixed temperature and total density. |
Some of the functions, like setState_TPX() have multiple forms depending upon the format for how the species compositions are set.
Molar Basis vs. Molality Basis
Mechanical properties
The ThermoPhase object specifies the mechanical equation of state of the phase. Functions which are defined at the ThermoPhase level to give the user more information about the mechanical properties are:
- ThermoPhase::pressure()
- ThermoPhase::isothermalCompressibility()
- ThermoPhase::thermalExpansionCoeff()
Treatment of the Phase Potential and the electrochemical potential of a species
The electrochemical potential of species k in a phase p, 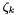 , is related to the chemical potential via the following equation,
, is related to the chemical potential via the following equation,
![\[ \zeta_{k}(T,P) = \mu_{k}(T,P) + z_k \phi_p \]](form_469.png)
where 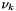 is the charge of species k, and
is the charge of species k, and 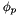 is the electric potential of phase p.
is the electric potential of phase p.
The potential 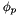 is tracked and internally storred within the base ThermoPhase object. It constitutes a specification of the internal state of the phase; it's the third state variable, the first two being temperature and density (or, pressure, for incompressible equations of state). It may be set with the function, ThermoPhase::setElectricPotential(), and may be queried with the function ThermoPhase::electricPotential().
is tracked and internally storred within the base ThermoPhase object. It constitutes a specification of the internal state of the phase; it's the third state variable, the first two being temperature and density (or, pressure, for incompressible equations of state). It may be set with the function, ThermoPhase::setElectricPotential(), and may be queried with the function ThermoPhase::electricPotential().
Note, the overall electrochemical potential of a phase may not be changed by the potential because many phases enforce charge neutrality:
![\[ 0 = \sum_k z_k X_k \]](form_472.png)
Whether charge neutrality is necessary for a phase is also specified within the ThermoPhase object, by the function call ThermoPhase::chargeNeutralityNecessary(). Note, that it is not necessary for the IdealGas phase, currently. However, it is necessary for liquid phases such as Cantera::DebyeHuckel and Cantera::HMWSoln for the proper specification of the chemical potentials.
This equation, when applied to the 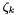 equation described above, results in a zero net change in the effective Gibbs free energy of the phase. However, specific charged species in the phase may increase or decrease their electochemical potentials, which will have an effect on interfacial reactions involving charged species, when there is a potential drop between phases. This effect is used within the Cantera::InterfaceKinetics and Cantera::EdgeKinetics kinetics objects classes.
equation described above, results in a zero net change in the effective Gibbs free energy of the phase. However, specific charged species in the phase may increase or decrease their electochemical potentials, which will have an effect on interfacial reactions involving charged species, when there is a potential drop between phases. This effect is used within the Cantera::InterfaceKinetics and Cantera::EdgeKinetics kinetics objects classes.
Other internal state variables, that track the treatment of other potential energy contributions, by adding contributions to the chemical potential to create an effective chemical potential, may be added at a later time.
Specification of Activities and Activity Conventions
The activity 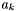 and activity coefficient
and activity coefficient 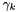 of a species in solution is related to the chemical potential by
of a species in solution is related to the chemical potential by
![\[ \mu_k = \mu_k^0(T,P) + \hat R T \log a_k.= \mu_k^0(T,P) + \hat R T \log x_k \gamma_k \]](form_473.png)
The quantity 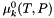 is the standard chemical potential at unit activity, which depends on the temperature and pressure, but not on the composition. The activity is dimensionless. Within liquid electrolytes its common to use a molality convention, where solute species employ the molality-based activity coefficients:
is the standard chemical potential at unit activity, which depends on the temperature and pressure, but not on the composition. The activity is dimensionless. Within liquid electrolytes its common to use a molality convention, where solute species employ the molality-based activity coefficients:
![\[ \mu_k = \mu_k^\triangle(T,P) + R T ln(a_k^{\triangle}) = \mu_k^\triangle(T,P) + R T ln(\gamma_k^{\triangle} \frac{m_k}{m^\triangle}) \]](form_474.png)
And, the solvent employs the following convention
![\[ \mu_o = \mu^o_o(T,P) + RT ln(a_o) \]](form_31.png)
where 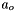 is often redefined in terms of the osmotic coefficient
is often redefined in terms of the osmotic coefficient 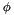 .
.
![\[ \phi = \frac{- ln(a_o)}{\tilde{M}_o \sum_{i \ne o} m_i} \]](form_363.png)
ThermoPhase classes which employ the molality based convention are all derived from the MolalityVPSSTP class. See the class description for further information on its capabilities.
The activity convention used by a ThermoPhase object may be queried via the ThermoPhase::activityConvention() function. A zero means molar based, while a one means molality based.
The function ThermoPhase::getActivities() returns a vector of activities. Whether these are molar-based or molality-based depends on the value of activityConvention().
The function getActivityCoefficients() always returns molar-based activity coefficients regardless of the activity convention used. The function MolalityVPSSTP::getMolalityActivityCoefficients() returns molality based activity coefficients for those ThermoPhase objects derived from the MolalityVPSSTP class. The function MolalityVPSSTP::osmoticCoefficient() returns the osmotic coefficient.
Activity Concentrations: Relationship of ThermoPhase to Kinetics Expressions
Cantera can handle both thermodynamics and kinetics mechanisms. Reversible kinetics mechanisms within Cantera must be compatible with thermodynamics in the sense that at equilibrium, or at infinite times, the concentrations of species must conform to thermodynamics. This means that for every valid reversible kinetics reaction in a mechanism, it must be reducible to an expression involving the ratio of the product activity to the reactant activities being equal to the exponential of the dimensionless standard state gibbs free energies of reaction. Irreversible kinetics reactions do not have this requirement; however, their usage can yield unexpected and inconsistent results in many situations. The actual units used in a kinetics expression depend on the context or the relative field of study. For example, in gas phase kinetics, species in kinetics expressions are expressed in terms of concentrations, i.e., gmol cm-3. In solid phase studies, however, kinetics is usually expressed in terms of unitless activities, which most often equate to solid phase mole fractions. In order to accomodate variability here, Cantera has come up with the idea of activity concentrations, 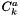 . Activity concentrations are the expressions used directly in kinetics expressions. These activity (or generalized) concentrations are used by kinetics manager classes to compute the forward and reverse rates of elementary reactions. Note that they may or may not have units of concentration --- they might be partial pressures, mole fractions, or surface coverages, The activity concentrations for species k,
. Activity concentrations are the expressions used directly in kinetics expressions. These activity (or generalized) concentrations are used by kinetics manager classes to compute the forward and reverse rates of elementary reactions. Note that they may or may not have units of concentration --- they might be partial pressures, mole fractions, or surface coverages, The activity concentrations for species k, 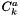 , are related to the activity for species, k,
, are related to the activity for species, k, 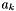 , via the following expression:
, via the following expression:
![\[ a_k = C^a_k / C^0_k \]](form_475.png)
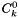 are called standard concentrations. They serve as multiplicative factors bewteen the activities and the generalized concentrations. Standard concentrations may be different for each species. They may depend on both the temperature and the pressure. However, they may not depend on the composition of the phase. For example, for the IdealGasPhase object the standard concentration is defined as
are called standard concentrations. They serve as multiplicative factors bewteen the activities and the generalized concentrations. Standard concentrations may be different for each species. They may depend on both the temperature and the pressure. However, they may not depend on the composition of the phase. For example, for the IdealGasPhase object the standard concentration is defined as
![\[ C^0_k = P/ R T \]](form_476.png)
In many solid phase kinetics problems,
![\[ C^0_k = 1.0 , \]](form_477.png)
is employed making the units for activity concentrations in solids unitless.
ThermoPhase member functions dealing with this concept include ThermoPhase::getActivityConcentrations() , which provides a vector of the current activity concentrations. The function, ThermoPhase::standardConcentration(int k=0) returns the standard concentration of the kth species. The function, ThermoPhase::logStandardConc(int k=0), returns the natural log of the kth standard concentration. The function ThermoPhase::getUnitsStandardConc() returns a vector of doubles, specifying the MKS units of the standard concentration of the kth species.
Initialization of ThermoPhase Objects within Cantera
Instantiation of ThermoPhase properties occurs by reading and processing the XML data contained within an ctxml data file. First a call to newPhase(std::string file, std::string id) or newPhase(XML_Node &phase) is made. The arguments serve to specify the XML data structure containing the phase information.
Within newPhase() a determination of what type of ThermoPhase object should be used is made. This is done within the routine ThermoFactory::newThermoPhase(std::string model) or related routines. Once the correct ThermoPhase derived object is selected and instantiated with a bare constructor, the function Cantera::importPhase() is called with the ThermoPhase derived object as one of its arguments.
Within importPhase(), a decision is made as to what type of standard state, i.e., either a reference state (just T dependent) or a standard state (both P and T dependent), is to be used to calculate the standard state properties of the species within the phase. If only a reference state is needed then a call to newSpeciesThermoMgr() is made in order pick a manager, i.e., a derivative of the SpeciesThermo object, to use.
If a temperature and pressure dependent standard state is needed then a call to VPSSMgrFactory::newVPSSMgr() is made in order pick a manager, i.e., a derivative of the VPSSMgr object, to use. Along with the VPSSMgr designation comes a determination of whether there is an accompanying SpeciesThermo and what type of SpeciesThermo object to use in the VPSSMgr calculations.
Once these determinations are made, the ThermoPhase object is ready to start reading in the species information, which includes all of the available standard state information about the species. this is done within the routine installSpecies().
Within installSpecies(), most of the common steps for adding a species are carried out. The element stoichiometry is read and elements are added as needed to the list of elements kept with the ThermoPhase object. The charge of the species is read in. The species is added into the list of species kept within the ThermoPhase object. Lastly, the standard state thermodynamics for the species is read in. For reference states, the routine, SpeciesThermoFactory::installThermoForSpecies(), is used to read in the data. Essentially, this routine is a factory routine for picking the correct subroutine to call to read the XML data from the input file and install the correct SpeciesThermoInterpType object into the SpeciesThermo object.
Within installSpecies(), for standard states, the routine, SpeciesThermoFactory::installVPThermoForSpecies() is called. However, this is just a shell routine for calling the VPSSMgr's derived VPSSMgr::createInstallPDSS() routine. Within the VPSSMgr::createInstallPDSS() routine of the derived VPSSMgr's object, the XML data from the input file is read and the calculations for the species standard state is installed. Additionally, the derived PDSS object is created and installed into the VPStandardStateTP list containing all of the PDSS objects for that phase.
Now that all of the species standard states are read in and installed into the ThermoPhase object, control once again is returned to the importPhase() function. Two derived functions are then called. The first one, ThermoPhase::initThermo(), is called. In this routine, all internal arrays within the ThermoPhase object are dimensioned according to the number of elements and species. Then, the function ThermoPhase::initThermoXML() is called. This function is tasked with reading in all of the thermodynamic function information specific to the calculation of the phase information. This includes all of the information about the activity coefficient calculation.
After the ThermoPhase::initThermoXML() is finished, the ThermoPhase routine is ready to receive requests for thermodynamic property information.
There is an alternative way to instantiate ThermoPhase objects that is applicable to a significant proportion of ThermoPhase classes. The phase may be instantiated via a constructor that invokes the XML data structure wherein the phase information is to be read directly. In this case, the call to newPhase() and the call to ThermoFactory::newThermoPhase(std::string model) is not made. However, soon after that, the call to importPhase() is made and thereafter instantiation follows the initialization course described previously in order to avoid as much duplicate code as possible. This alternative way to instantiate ThermoPhase objects has the advantage of working well with hard-coded situations. And, it works well also with situations where new ThermoPhase classes are being developed and haven't yet made their way into the factory routines.
Adding Additional Thermodynamics Models
In general, factory routines throw specific errors when encountering unknown thermodynamics models in XML files. All of the error classes derive from the class, CanteraError. The newVPSSMgr() routines throws the UnknownVPSSMgr class error when they encounter an unknown string in the XML input file specifying the VPSSMgr class to use.
Many of the important member functions in factory routines are virtual classes. This means that a user may write their own factory classes which inherit from the base Cantera factory classes to provide additional ThermoPhase classes.
- See also:
- newPhase(std::string file, std::string id) Description for how to read ThermoPhases from XML files.
- newPhase(XML_Node &phase) How to call the Factory routine to create and initialize ThermoPhase objects.
Standard ThermoPhase objects may be instantiated by calling the main Cantera factory class for ThermoPhase objects; This class is called ThermoFactory.
Function Documentation
| std::string eosTypeString | ( | int | ieos, | |
| int | length = 100 | |||
| ) |
Translate the eosType id into a string.
Returns a string representation of the eosType id for a phase.
- Parameters:
-
ieos eosType id of the phase. This is unique for the phase length maximum length of the return string. Defaults to 100
- Returns:
- returns a string representation.
| bool importPhase | ( | XML_Node & | phase, | |
| ThermoPhase * | th, | |||
| SpeciesThermoFactory * | spfactory = 0 | |||
| ) |
Import a phase information into an empty thermophase object.
Here we read an XML description of the thermodynamic information for a phase. At the end of this routine, the phase should be ready to be used within applications. This routine contains some key routines that are used as pass back routines so that the phase (and the contents of the XML file) may contain variable paramerizations for the specification of the species standard states, the equation of state, and the specification of other nonidealities. Below, a description is presented of the main algorithm for bringing up a ThermoPhase object, with care to present points where customizations occur.
Before invoking this routine, either the ThermoPhase Factory routines are called or direct constructor routines are called that instantiate an inherited ThermoPhase object. This object is input to this routine, and therefore contains inherited routines that drive the custimation of the initialization process.
At the start of the routine, we import descriptions of the elements that make up the species in a phase.
We call setParametersFromXML(eos) to read parameters about the thermo phase before the species are read in.
We call addElementsFromXML() to add elements into the description of the phase.
We create a new species thermo manager. Function 'newSpeciesThermoMgr' looks at the species in the database to see what thermodynamic property parameterizations are used, and selects a class that can handle the parameterizations found.
We import information about the species, including their reference state thermodynamic polynomials. We then freeze the state of the species in the element.
Finally, we call initThermoXML(), a member function of the ThermoPhase object, to "finish" the description. Now that the species are known, additional information may be read in about the thermodynamics of the phase, (e.g., virial coefficients, which are binary or ternary interaction parameters between species).
- Parameters:
-
phase This object must be the phase node of a complete XML tree description of the phase, including all of the species data. In other words while "phase" must point to an XML phase object, it must have sibling nodes "speciesData" that describe the species in the phase. th Pointer to the ThermoPhase object which will handle the thermodynamics for this phase. We initialize part of the Thermophase object here, especially for those objects which are part of the Cantera Kernel. spfactory species Thermo factory pointer, if available. If not available, one will be created.
Referenced by WaterSSTP::constructPhaseXML(), IdealSolidSolnPhase::constructPhaseXML(), IdealMolalSoln::constructPhaseXML(), HMWSoln::constructPhaseXML(), DebyeHuckel::constructPhaseXML(), electrodeElectron::electrodeElectron(), MetalSHEelectrons::MetalSHEelectrons(), MineralEQ3::MineralEQ3(), StoichSubstanceSSTP::StoichSubstanceSSTP(), and SurfPhase::SurfPhase().
| bool installSpecies | ( | int | k, | |
| const XML_Node & | s, | |||
| thermo_t & | p, | |||
| SpeciesThermo * | spthermo_ptr, | |||
| int | rule, | |||
| XML_Node * | phaseNode_ptr = 0, |
|||
| VPSSMgr * | vpss_ptr = 0, |
|||
| SpeciesThermoFactory * | factory = 0 | |||
| ) |
Install a species into a ThermoPhase object, which defines the phase thermodynamics and speciation.
This routine first gathers the information from the Species XML tree and calls addUniqueSpecies() to add it to the ThermoPhase object, p. This information consists of: ecomp[] = element composition of species. chgr = electric charge of species name = string name of species sz = size of the species (option double used a lot in thermo)
Then, the routine processes the "thermo" XML element and calls underlying utility routines to read the XML elements containing the thermodynamic information for the reference state of the species. Failures or lack of information trigger an "UnknownSpeciesThermoModel" exception being thrown.
- Parameters:
-
k Species Index in the phase s XML_Node containing the species data for this species. p Reference to the ThermoPhase object. spthermo_ptr Reference to the SpeciesThermo object, where the standard state thermo properties for this species will be installed. rule Parameter that handles what to do with species who have elements that aren't declared. Check that all elements in the species exist in 'p'. If rule != 0, quietly skip this species if some elements are undeclared; otherwise, throw an exception phaseNode_ptr Pointer to the XML_Node for this phase (defaults to 0) vpss_ptr pointer to the Manager that calculates standard state thermo properties factory Pointer to the SpeciesThermoFactory . (defaults to 0)
- Returns:
- Returns true if everything is ok, false otherwise.
| ThermoPhase * newPhase | ( | std::string | infile, | |
| std::string | id | |||
| ) |
Create and Initialize a ThermoPhase object from an XML input file.
This routine is a wrapper around the newPhase(XML_Node) routine which does the work. The wrapper locates the input phase XML_Node in a file, and then instantiates the object, returning the pointer to the ThermoPhase object.
- Parameters:
-
infile name of the input file id name of the phase id in the file. If this is blank, the first phase in the file is used.
- Returns:
- Returns an initialized ThermoPhase object.
| ThermoPhase* Cantera::newThermoPhase | ( | std::string | model, | |
| ThermoFactory * | f = 0 | |||
| ) | [inline] |
Create a new thermo manager instance.
- Parameters:
-
model String to look up the model against f ThermoFactor instance to use in matching the string
- Returns:
- Returns a pointer to a new ThermoPhase instance matching the model string. Returns NULL if something went wrong. Throws an exception UnknownThermoPhaseModel if the string wasn't matched.
Definition at line 138 of file ThermoFactory.h.
References ThermoFactory::factory().
| const XML_Node * speciesXML_Node | ( | std::string | kname, | |
| const XML_Node * | phaseSpeciesData | |||
| ) |
Search an XML tree for species data.
This utility routine will search the XML tree for the species named by the string, kname. It will return the XML_Node pointer to the species data for that species. Failures of any kind return the null pointer.
- Parameters:
-
kname String containing the name of the species. phaseSpeciesData Pointer to the XML speciesData element containing the species data for that phase.
 1.6.3
1.6.3