Lab 1
In this lab, we’ll try to cover some of the basic ways of interacting with R and then pretty quickly switch to data wrangling and data visualization. I recommend using RStudio, https://support.rstudio.com, because they have lots of great resources (e.g., Help –> Cheatsheets).
Another great resource for many of the functions we’ll use: http://r4ds.had.co.nz/.
Calculator
First of all, R can be used as a calculator:
1 + 1## [1] 22 * 3## [1] 63 ^ 2## [1] 95 %% 3## [1] 2It’s a good idea not to work directly in the console but to have a script where you will first write your commands and then execute them in the console. Once you open a new R script there are a few useful things to note:
- Use
#to comment lines so they don’t get executed - You can send a line directly to console from script with
command + return(on mac) orctrl + enter(windows)
Variables
Values can be stored as variables
a <- 2
a## [1] 2b <- 2 + 2
b## [1] 4c <- b + 3
c## [1] 7When you’re defining variables, try to use meaningful names like average_of_ratings rather than variable1.
Functions
A function is an object that takes some arguments and returns a value.
log(4)## [1] 1.386294print("hello world")## [1] "hello world"help(log)
?logVectors
If you want to store more than one thing in a variable you may want to make a vector.To do this you will use the function c() which combines values
v1 <- c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10)
v1## [1] 1 2 3 4 5 6 7 8 9 10HINT: If you are ever unsure about how a function works or what arguments it takes. Typing ?[FUNCTION NAME GOES HERE] will open up a help file (e.g., ?c).
Now that we have some values stored in a vector, we may want to access those values and we can do this by using their position in the vector or index.
v1[1]## [1] 1v1[5]## [1] 5v1[-1]## [1] 2 3 4 5 6 7 8 9 10v1[c(2, 7)]## [1] 2 7v1[-c(2, 7)]## [1] 1 3 4 5 6 8 9 10v1[1:3]## [1] 1 2 3v1[-(1:3)]## [1] 4 5 6 7 8 9 10You may also want to get some overall information about the vector:
what types of values are in
v1?str(v1)## num [1:10] 1 2 3 4 5 6 7 8 9 10summary(v1)## Min. 1st Qu. Median Mean 3rd Qu. Max. ## 1.00 3.25 5.50 5.50 7.75 10.00how long is
v1?length(v1)## [1] 10what is the average of all these values?
mean(v1)## [1] 5.5what is the standard deviation of all these values?
sd(v1)## [1] 3.02765
Earlier we created v1 by simply listing all the elements in it and usingc() but if you have lots of values, this is very tedious. There are some functions that can help make vectors more efficiently
v2 <- (1:10)
v2## [1] 1 2 3 4 5 6 7 8 9 10v3 <- rep(x = 1, times = 10)
v3## [1] 1 1 1 1 1 1 1 1 1 1v4 <- rep(1:2, 5)
v4## [1] 1 2 1 2 1 2 1 2 1 2v5 <- seq(from = 1, to = 20, by = 2)
v5## [1] 1 3 5 7 9 11 13 15 17 19Note that for v4, I didn’t include the names of the arguments but R figures out which is which by the order
You can also apply operations to all elements of the vector simultaneously.
v1 + 1## [1] 2 3 4 5 6 7 8 9 10 11v1 * 100## [1] 100 200 300 400 500 600 700 800 900 1000You can also do pair-wise operations on 2 vectors.
v1 + v2## [1] 2 4 6 8 10 12 14 16 18 20Characters
So far we’ve looked at numeric variables and vectors, but they can also be strings
name <- "Rachel"
name## [1] "Rachel"friends <- c("Rachel", "Ross", "Joey", "Monica", "Chandler", "Phoebe")
friends## [1] "Rachel" "Ross" "Joey" "Monica" "Chandler" "Phoebe"str(friends)## chr [1:6] "Rachel" "Ross" "Joey" "Monica" "Chandler" "Phoebe"You can even store numbers as strings (and sometimes data you load from a file will be stored this way so watch out for that)
some_numbers <- c("2", "3", "4")…but you can’t manipulate them as numbers
some_numbers + 1So you might want to convert the strings into numbers first using as.numeric()
some_numbers <- as.numeric(some_numbers)
some_numbers + 1## [1] 3 4 5NA
Another important datatype is NA. Say I’m storing people’s heights in inches in a dataframe, but I don’t have data on the third person.
heights <- c(72, 70, NA, 64)
str(heights)## num [1:4] 72 70 NA 64Even though it’s composed of letters, NA is not a string, in this case it’s numeric, and represents a missing value, or an invalid value, or whatever. You can still perform operations on the height vector:
heights + 1## [1] 73 71 NA 65heights * 2## [1] 144 140 NA 128if you had an NA in a vector of strings, its datatype would be a character.
friends <- c("Rachel", NA, "Joey", "Monica", "Chandler", "Phoebe")
str(friends)## chr [1:6] "Rachel" NA "Joey" "Monica" "Chandler" "Phoebe"If you have NA in your vector and want to use a function on it, this can complicate things
mean(heights)## [1] NATo avoid returning NA, you may want to just throw out the NA values using na.omit and work with what’s left.
heights_no_NA <- na.omit(heights)
mean(heights_no_NA)## [1] 68.66667Alternatively, many functions have a built-in argument na.rm that you can use to tell the function what to do about NA values. So you can do the previous step in 1 line:
mean(heights_no_NA, na.rm = TRUE)## [1] 68.66667It can also be useful to know if a vector contains NA ahead of time and where those values are:
is.na(heights)## [1] FALSE FALSE TRUE FALSEwhich(is.na(heights))## [1] 3Booleans
This brings us to another important datatype: booleans. They are TRUE or FALSE, or T or F. Here are some expressions that return boolean values:
1 < 100## [1] TRUE500 == 500 # for equality testing, use double-equals!## [1] TRUE1 == 2 | 2 == 2 # OR## [1] TRUE1 == 1 & 100 == 100 # AND## [1] TRUE1 == 1 & 100 == 101 # AND## [1] FALSETry it yourself…
Make a vector, “tens” of all the multiples of 10 up to 200.
Find the indices of the numbers divisible by 3
tens <- seq(from = 10, to = 200, by = 10) tens## [1] 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 ## [18] 180 190 200which(tens %% 3 == 0)## [1] 3 6 9 12 15 18
Dataframes
Most data you will work with in R will be in a dataframe format. Dataframes are basically tables where each column is a vector with a name. Dataframes can combine character vectors, numeric vectors, logical (boolean) vectors, etc. This is really useful for data from experiments where you may want one column to contain information about the name of the condition (a string) and another column to contain response times (a number).
Let’s read in some data!
But first a digression… One of the best things about R is that it is open-source and lots of R users who find that some functionality is missing from base R (which is what we’ve been using so far) will write their own functions and then share them with the R community. Often times they’ll write whole packages of functions to greatly enhance the capabilities of base R. In order for you to use those packages, they need to be installed on your computer and loaded up in your current session. For current purposes, you will need the tidyverse package and you can install it with this simple command:
install.packages("tidyverse")When the installation is done, load up the library of functions in the package with the following command:
library(tidyverse)
library(stringr)Okay, digression over.
Let’s read in your lexical decision data from earlier using a tidyverse function called read_csv().
data_source <- "http://web.mit.edu/psycholinglab/data/"
rt_data <- read_csv(file.path(data_source, "in_lab_rts_2018.csv"))Our data is now stored as a dataframe. The output message tells us what datatype read_csv() assigned to every column. It usually does a pretty good job of guessing the appropriate datatype but on occasion you may have to correct using a function like as.numeric() or as.character().
Note that the path to the data can be any folder on your computer or online (a url). If you just put in the filename without the path, it will look for the file in the local folder.
Also note, that if you have column headers in your csv file, read_csv() will automatically name your columns accordingly and you won’t have to specify col_names=.
At this point, it’s a good idea to look at your data to make sure everything was correctly uploaded. In R Studio, you can open up a viewing pane with the command View(d) to see the data in spreadsheet form. You can also use summary(), str() and glimpse()
glimpse(rt_data)## Observations: 264
## Variables: 5
## $ time <dbl> 1.518185e+12, 1.518185e+12, 1.518185e+12, 1.518185e+12...
## $ subject <int> 9, 9, 7, 9, 7, 9, 8, 6, 3, 7, 9, 0, 8, 6, 3, 9, 7, 8, ...
## $ word <chr> "book", "noosin", "book", "eat", "noosin", "goamboozle...
## $ trial <int> 0, 1, 0, 2, 1, 3, 0, 0, 0, 2, 4, 0, 1, 1, 1, 5, 3, 2, ...
## $ rt <dbl> 2.2346671, 0.8533659, 3.4355321, 0.5956440, 0.8158729,...You can also extract just the names of the columns:
names(rt_data)## [1] "time" "subject" "word" "trial" "rt"Or just the first (or last) few rows of the dataframe:
head(rt_data)## # A tibble: 6 x 5
## time subject word trial rt
## <dbl> <int> <chr> <int> <dbl>
## 1 1.518185e+12 9 book 0 2.2346671
## 2 1.518185e+12 9 noosin 1 0.8533659
## 3 1.518185e+12 7 book 0 3.4355321
## 4 1.518185e+12 9 eat 2 0.5956440
## 5 1.518185e+12 7 noosin 1 0.8158729
## 6 1.518185e+12 9 goamboozle 3 0.6285422tail(rt_data)## # A tibble: 6 x 5
## time subject word trial rt
## <dbl> <int> <chr> <int> <dbl>
## 1 1.518185e+12 14 seefer 18 0.7138901
## 2 1.518185e+12 14 sqw 19 0.6964531
## 3 1.518185e+12 13 encyclopedia 20 1.5294392
## 4 1.518185e+12 14 encyclopedia 20 0.9169972
## 5 1.518185e+12 13 understandable 21 1.0834951
## 6 1.518185e+12 14 understandable 21 0.5998409Or look at the dimensions of your dataframe
dim(rt_data)## [1] 264 5nrow(rt_data)## [1] 264ncol(rt_data)## [1] 5To access a specific column, row, or cell you can use indexing in much the same way you can with vectors (just now with 2 dimensions)
rt_data[1, 2] # what's in the 1st row, 2nd column## # A tibble: 1 x 1
## subject
## <int>
## 1 9rt_data[1, ] # the 1st row for all columns## # A tibble: 1 x 5
## time subject word trial rt
## <dbl> <int> <chr> <int> <dbl>
## 1 1.518185e+12 9 book 0 2.234667rt_data[, 2] # all rows for the 2nd column## # A tibble: 264 x 1
## subject
## <int>
## 1 9
## 2 9
## 3 7
## 4 9
## 5 7
## 6 9
## 7 8
## 8 6
## 9 3
## 10 7
## # ... with 254 more rowsrt_data[, c(2, 5)] # all rows for columns 2 and 5## # A tibble: 264 x 2
## subject rt
## <int> <dbl>
## 1 9 2.2346671
## 2 9 0.8533659
## 3 7 3.4355321
## 4 9 0.5956440
## 5 7 0.8158729
## 6 9 0.6285422
## 7 8 0.7227321
## 8 6 1.2849419
## 9 3 1.0876911
## 10 7 0.6137280
## # ... with 254 more rowsrt_data[, c("subject", "rt")]## # A tibble: 264 x 2
## subject rt
## <int> <dbl>
## 1 9 2.2346671
## 2 9 0.8533659
## 3 7 3.4355321
## 4 9 0.5956440
## 5 7 0.8158729
## 6 9 0.6285422
## 7 8 0.7227321
## 8 6 1.2849419
## 9 3 1.0876911
## 10 7 0.6137280
## # ... with 254 more rowsAnother easy way to extract a dataframe column is by using the $ operator and the column name
head(rt_data$rt)## [1] 2.2346671 0.8533659 3.4355321 0.5956440 0.8158729 0.6285422rt_data$rt is a (numeric) vector so you can perform various operations on it (as we saw earlier)
head(rt_data$rt * 2)## [1] 4.469334 1.706732 6.871064 1.191288 1.631746 1.257084mean(rt_data$rt)## [1] 0.866832Data manipulation (dplyr)
Often when you upload data it’s not yet in a convenient, “tidy” form so data wrangling refers to the various cleaning and re-arranging steps between uploading data and being able to visualize or analyze it. I’ll start out by showing you a few of the most common things you might want to do with your data.
For example, in this dataset, we want to know about how response times on the lexical decision task might differ depending on whether it’s a real word or a non-word but this information is missing from our data.
Let’s see what words were included in the experiment.
words <- unique(rt_data$word)
words## [1] "book" "noosin" "eat"
## [4] "goamboozle" "condition" "xdqww"
## [7] "word" "retire" "feffer"
## [10] "fly" "qqqwqw" "coat"
## [13] "condensationatee" "sporm" "art"
## [16] "goam" "gold" "three"
## [19] "seefer" "sqw" "encyclopedia"
## [22] "understandable"Now let’s make a vector containing only the real words:
real_words <- words[c(1, 3, 5, 7, 8, 10, 12, 15, 17, 18, 21, 22)]
real_words## [1] "book" "eat" "condition" "word"
## [5] "retire" "fly" "coat" "art"
## [9] "gold" "three" "encyclopedia" "understandable"We can check if a value is represented in an array using the operator %in%.
"cat" %in% c("cat", "dog", "horse")## [1] TRUEc("cat", "ocelot") %in% c("cat", "dog", "horse")## [1] TRUE FALSEmutate
Now we can add a column to our dataframe, rt_data, that contains that condition information. We’re going to do this using the mutate() function.
rt_data <- mutate(rt_data, is_real = word %in% real_words)
head(rt_data)## # A tibble: 6 x 6
## time subject word trial rt is_real
## <dbl> <int> <chr> <int> <dbl> <lgl>
## 1 1.518185e+12 9 book 0 2.2346671 TRUE
## 2 1.518185e+12 9 noosin 1 0.8533659 FALSE
## 3 1.518185e+12 7 book 0 3.4355321 TRUE
## 4 1.518185e+12 9 eat 2 0.5956440 TRUE
## 5 1.518185e+12 7 noosin 1 0.8158729 FALSE
## 6 1.518185e+12 9 goamboozle 3 0.6285422 FALSEmutate() is extremely useful anytime you want to add information to your data. For instance, the reaction times here appear to be in seconds but maybe we want to look at them in milliseconds. Maybe we also want to code if the word starts with the letter “b” and code which words are longer than 6 letters long. This can be done all at once.
mutate(rt_data,
rt_ms = rt * 1000,
starts_d = str_sub(word, 1, 1) == "b",
longer_than_6 = if_else(str_length(word) > 6, "long", "short"))## # A tibble: 264 x 9
## time subject word trial rt is_real rt_ms
## <dbl> <int> <chr> <int> <dbl> <lgl> <dbl>
## 1 1.518185e+12 9 book 0 2.2346671 TRUE 2234.6671
## 2 1.518185e+12 9 noosin 1 0.8533659 FALSE 853.3659
## 3 1.518185e+12 7 book 0 3.4355321 TRUE 3435.5321
## 4 1.518185e+12 9 eat 2 0.5956440 TRUE 595.6440
## 5 1.518185e+12 7 noosin 1 0.8158729 FALSE 815.8729
## 6 1.518185e+12 9 goamboozle 3 0.6285422 FALSE 628.5422
## 7 1.518185e+12 8 book 0 0.7227321 TRUE 722.7321
## 8 1.518185e+12 6 book 0 1.2849419 TRUE 1284.9419
## 9 1.518185e+12 3 book 0 1.0876911 TRUE 1087.6911
## 10 1.518185e+12 7 eat 2 0.6137280 TRUE 613.7280
## # ... with 254 more rows, and 2 more variables: starts_d <lgl>,
## # longer_than_6 <chr>Note: + str_sub() extracts a subset of word starting at position 1 and ending at position 1 (i.e., just the first letter). + if_else() is a useful function which takes a logical comparison as a first argument and then what to do if it is TRUE as the second argument and what to do if it is FALSE as the third.
filter
Now let’s say we want to look at only a subset of the data, we can filter() it:
filter(rt_data, str_length(word) > 6)## # A tibble: 60 x 6
## time subject word trial rt is_real
## <dbl> <int> <chr> <int> <dbl> <lgl>
## 1 1.518185e+12 9 goamboozle 3 0.6285422 FALSE
## 2 1.518185e+12 9 condition 4 0.6441879 TRUE
## 3 1.518185e+12 7 goamboozle 3 1.2371540 FALSE
## 4 1.518185e+12 7 condition 4 0.7699041 TRUE
## 5 1.518185e+12 8 goamboozle 3 0.6066039 FALSE
## 6 1.518185e+12 6 goamboozle 3 0.6936800 FALSE
## 7 1.518185e+12 3 goamboozle 3 0.7247601 FALSE
## 8 1.518185e+12 8 condition 4 0.7711821 TRUE
## 9 1.518185e+12 6 condition 4 0.5985160 TRUE
## 10 1.518185e+12 0 goamboozle 3 0.8194630 FALSE
## # ... with 50 more rowsselect
If your dataframe is getting unruly, you can focus on a few key columns with select()
select(rt_data, subject, word, rt)## # A tibble: 264 x 3
## subject word rt
## <int> <chr> <dbl>
## 1 9 book 2.2346671
## 2 9 noosin 0.8533659
## 3 7 book 3.4355321
## 4 9 eat 0.5956440
## 5 7 noosin 0.8158729
## 6 9 goamboozle 0.6285422
## 7 8 book 0.7227321
## 8 6 book 1.2849419
## 9 3 book 1.0876911
## 10 7 eat 0.6137280
## # ... with 254 more rowsarrange
You can also sort the dataframe by one of the columns:
arrange(rt_data, rt)## # A tibble: 264 x 6
## time subject word trial rt is_real
## <dbl> <int> <chr> <int> <dbl> <lgl>
## 1 1.518185e+12 1 retire 7 0.0003647804 TRUE
## 2 1.518185e+12 1 condensationatee 12 0.0007541180 FALSE
## 3 1.518185e+12 13 three 17 0.3730161190 TRUE
## 4 1.518185e+12 13 art 14 0.4536800385 TRUE
## 5 1.518185e+12 8 word 6 0.4927048683 TRUE
## 6 1.518185e+12 13 sqw 19 0.4932930470 FALSE
## 7 1.518185e+12 14 coat 11 0.4945719242 TRUE
## 8 1.518185e+12 14 three 17 0.5010519028 TRUE
## 9 1.518185e+12 9 xdqww 5 0.5207970142 FALSE
## 10 1.518185e+12 0 word 6 0.5246582031 TRUE
## # ... with 254 more rowsgroup_by and summarise
Most of the time when you have data, the ultimate goal is to summarize it in some way. For example, you may want to know the mean response time for each subject by type of word (real vs. fake).
summarise(group_by(rt_data, subject, is_real), mean_rt = mean(rt))## # A tibble: 24 x 3
## # Groups: subject [?]
## subject is_real mean_rt
## <int> <lgl> <dbl>
## 1 0 FALSE 0.8384297
## 2 0 TRUE 0.8294404
## 3 1 FALSE 0.7580644
## 4 1 TRUE 0.8128119
## 5 3 FALSE 0.9057278
## 6 3 TRUE 0.8010959
## 7 4 FALSE 0.8137079
## 8 4 TRUE 0.6964481
## 9 6 FALSE 0.8390218
## 10 6 TRUE 0.7740091
## # ... with 14 more rowsAs you can see, we often want to string tidyverse functions together which can get difficult to read. The solution to this is…
%>%
We can create a pipeline where the dataframe undergoes various transformations one after the other with the same functions, mutate(), filter(), etc. without having to repeat the name of the dataframe over and over and much more intuitive syntax.
# this is the previous syntax
mutate(rt_data,
"rt_ms" = rt * 1000,
"starts_d" = str_sub(word, 1, 1) == "b",
"longer_than_6" = if_else(str_length(word) > 6, "long", "short"))## # A tibble: 264 x 9
## time subject word trial rt is_real rt_ms
## <dbl> <int> <chr> <int> <dbl> <lgl> <dbl>
## 1 1.518185e+12 9 book 0 2.2346671 TRUE 2234.6671
## 2 1.518185e+12 9 noosin 1 0.8533659 FALSE 853.3659
## 3 1.518185e+12 7 book 0 3.4355321 TRUE 3435.5321
## 4 1.518185e+12 9 eat 2 0.5956440 TRUE 595.6440
## 5 1.518185e+12 7 noosin 1 0.8158729 FALSE 815.8729
## 6 1.518185e+12 9 goamboozle 3 0.6285422 FALSE 628.5422
## 7 1.518185e+12 8 book 0 0.7227321 TRUE 722.7321
## 8 1.518185e+12 6 book 0 1.2849419 TRUE 1284.9419
## 9 1.518185e+12 3 book 0 1.0876911 TRUE 1087.6911
## 10 1.518185e+12 7 eat 2 0.6137280 TRUE 613.7280
## # ... with 254 more rows, and 2 more variables: starts_d <lgl>,
## # longer_than_6 <chr># this is the piping syntax
rt_data %>%
mutate("rt_ms" = rt * 1000,
"starts_d" = str_sub(word, 1, 1) == "b",
"longer_than_6" = if_else(str_length(word) > 6, "long", "short"))## # A tibble: 264 x 9
## time subject word trial rt is_real rt_ms
## <dbl> <int> <chr> <int> <dbl> <lgl> <dbl>
## 1 1.518185e+12 9 book 0 2.2346671 TRUE 2234.6671
## 2 1.518185e+12 9 noosin 1 0.8533659 FALSE 853.3659
## 3 1.518185e+12 7 book 0 3.4355321 TRUE 3435.5321
## 4 1.518185e+12 9 eat 2 0.5956440 TRUE 595.6440
## 5 1.518185e+12 7 noosin 1 0.8158729 FALSE 815.8729
## 6 1.518185e+12 9 goamboozle 3 0.6285422 FALSE 628.5422
## 7 1.518185e+12 8 book 0 0.7227321 TRUE 722.7321
## 8 1.518185e+12 6 book 0 1.2849419 TRUE 1284.9419
## 9 1.518185e+12 3 book 0 1.0876911 TRUE 1087.6911
## 10 1.518185e+12 7 eat 2 0.6137280 TRUE 613.7280
## # ... with 254 more rows, and 2 more variables: starts_d <lgl>,
## # longer_than_6 <chr>And we can keep adding functions to the pipeline very easily…
rt_data %>%
mutate("rt_ms" = rt * 1000,
"starts_d" = str_sub(word, 1, 1) == "b",
"longer_than_6" = if_else(str_length(word) > 6, "long", "short")) %>%
filter(rt > 0.002)## # A tibble: 262 x 9
## time subject word trial rt is_real rt_ms
## <dbl> <int> <chr> <int> <dbl> <lgl> <dbl>
## 1 1.518185e+12 9 book 0 2.2346671 TRUE 2234.6671
## 2 1.518185e+12 9 noosin 1 0.8533659 FALSE 853.3659
## 3 1.518185e+12 7 book 0 3.4355321 TRUE 3435.5321
## 4 1.518185e+12 9 eat 2 0.5956440 TRUE 595.6440
## 5 1.518185e+12 7 noosin 1 0.8158729 FALSE 815.8729
## 6 1.518185e+12 9 goamboozle 3 0.6285422 FALSE 628.5422
## 7 1.518185e+12 8 book 0 0.7227321 TRUE 722.7321
## 8 1.518185e+12 6 book 0 1.2849419 TRUE 1284.9419
## 9 1.518185e+12 3 book 0 1.0876911 TRUE 1087.6911
## 10 1.518185e+12 7 eat 2 0.6137280 TRUE 613.7280
## # ... with 252 more rows, and 2 more variables: starts_d <lgl>,
## # longer_than_6 <chr>rt_data %>%
mutate("rt_ms" = rt * 1000,
"starts_d" = str_sub(word, 1, 1) == "b",
"longer_than_6" = if_else(str_length(word) > 6, "long", "short")) %>%
filter(rt > 0.002) %>%
group_by(subject, is_real) %>%
summarise(mean_rt = mean(rt))## # A tibble: 24 x 3
## # Groups: subject [?]
## subject is_real mean_rt
## <int> <lgl> <dbl>
## 1 0 FALSE 0.8384297
## 2 0 TRUE 0.8294404
## 3 1 FALSE 0.8422099
## 4 1 TRUE 0.8866708
## 5 3 FALSE 0.9057278
## 6 3 TRUE 0.8010959
## 7 4 FALSE 0.8137079
## 8 4 TRUE 0.6964481
## 9 6 FALSE 0.8390218
## 10 6 TRUE 0.7740091
## # ... with 14 more rowsWe can look just at conditions and add some summary stats
rt_data %>%
group_by(is_real) %>%
summarise(mean_rt = mean(rt),
median_rt = median(rt),
sd_rt = sd(rt))## # A tibble: 2 x 4
## is_real mean_rt median_rt sd_rt
## <lgl> <dbl> <dbl> <dbl>
## 1 FALSE 0.9442195 0.8029714 0.4748030
## 2 TRUE 0.8023425 0.7043273 0.3776124It looks like average response time was longer for fake words.
Try it yourself…
Add a new column to
rt_datathat codes whether the word ends with “n”Get the means and counts for real and fake words split by whether they end in “n” or not
rt_data_n <- rt_data %>% mutate("ends_with_n" = str_sub(word, -1, -1) == "n") %>% group_by(is_real, ends_with_n) %>% summarize(mean_rt = mean(rt), n_rt = n()) rt_data_n## # A tibble: 4 x 4 ## # Groups: is_real [?] ## is_real ends_with_n mean_rt n_rt ## <lgl> <lgl> <dbl> <int> ## 1 FALSE FALSE 0.9212505 108 ## 2 FALSE TRUE 1.1509408 12 ## 3 TRUE FALSE 0.7912822 132 ## 4 TRUE TRUE 0.9240052 12
Data visualization (ggplot2)
Looking at columns of numbers isn’t really the best way to do data analysis. You could be tripped up by placement of decimal points, you might accidentally miss a big number. It would be much better if we could PLOT these numbers so we can visually tell if anything stands out.
If you’ve taken an introductory Psych or Neuro course you might know that a huge proportion of human cortex is devoted to visual information processing; we have hugely powerful abilities to process visual data. By using plotting we can leverage that ability to get a fast sense of what is going on in our data.
Visualizing your data is an extremely important part of any data analysis. tidyverse contains a whole library of functions for plotting: ggplot2. I’ll be showing you how to use these functions but also I’ll be trying to give you some intuitions about how researchers use visualization to get a better understanding of their data.
The first thing you might want to know is what your dependent variable, in this case the response time, looks like. In other words, how is it distributed?
Histograms
ggplot(data = rt_data) +
geom_histogram(mapping = aes(x = rt))
ggplot syntax might seem a little unusual. You can think of it as first creating a plot coordinate system with ggplot() and the you can add layers of information with +.
ggplot(data = rt_data) would create an empty plot because you haven’t told it anything about what variables you’re interested in or how you want to look at them. This is where geometric objects, or “geoms”, come in.
geom_histogram() is going to make this plot a histogram. A histogram has values of whatever variable you choose on the x axis and counts of those values on the y. The aesthetic mapping, aes(), arguments let us specify which variable, in this case rt, we want to know the distribution of. We can also change visual aspects of the geom, like the width of the bins, depending on what will make the graph more clear and informative.
ggplot(rt_data) +
geom_histogram(aes(x = rt), bins = 60)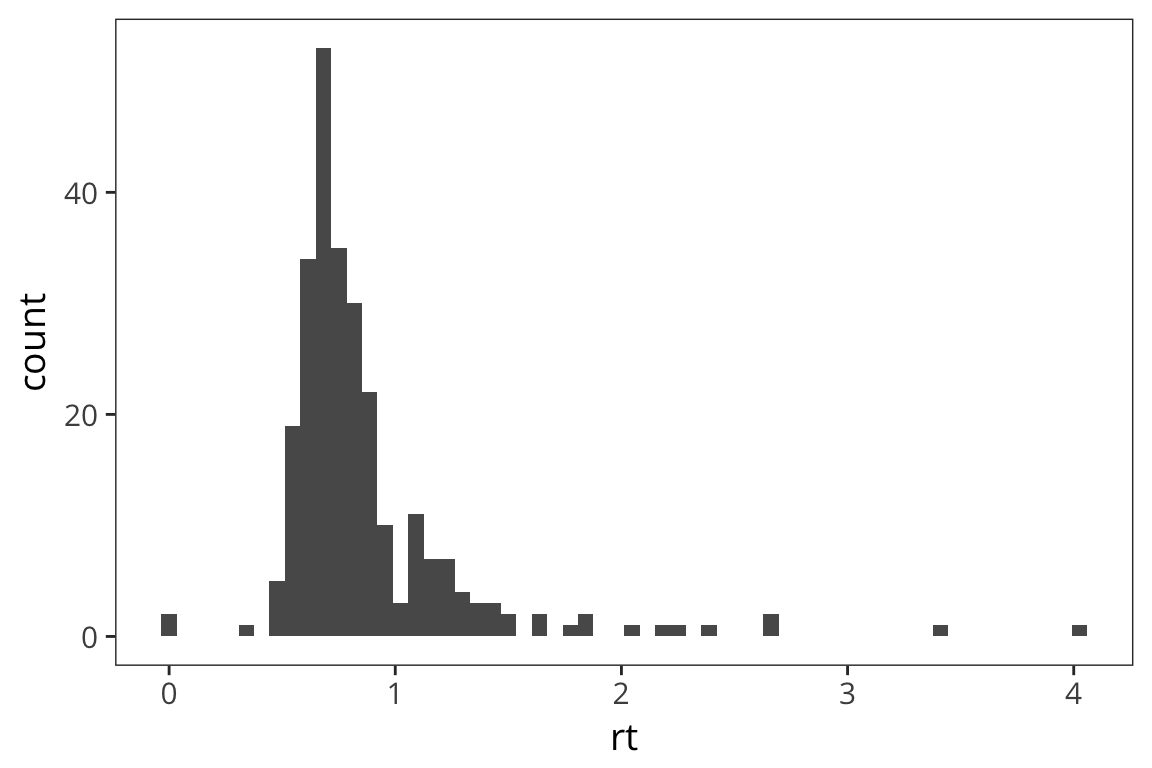
What can we learn from this histogram?
In this histogram we can see that there are a lot of response times around 1 second and a few longer outlier response times. This is important to know for when we analyze the response times because certain descriptive statistics like the mean are very sensitive to outliers.
Scatterplots
Let’s say we are curious to see if people speed up or slow down over the course of doing the lexical decision task. So, we want to plot trial number and compare it with mean rt. Let’s put trial number on the x axis, mean rt on the y axis, and make a scatterplot. For this we’re going to use a different geom, geom_point(). Contrary to geom_histogram() this takes a minimum of 2 arguments, x and y.
ggplot(rt_data) +
geom_point(aes(x = trial, y = rt))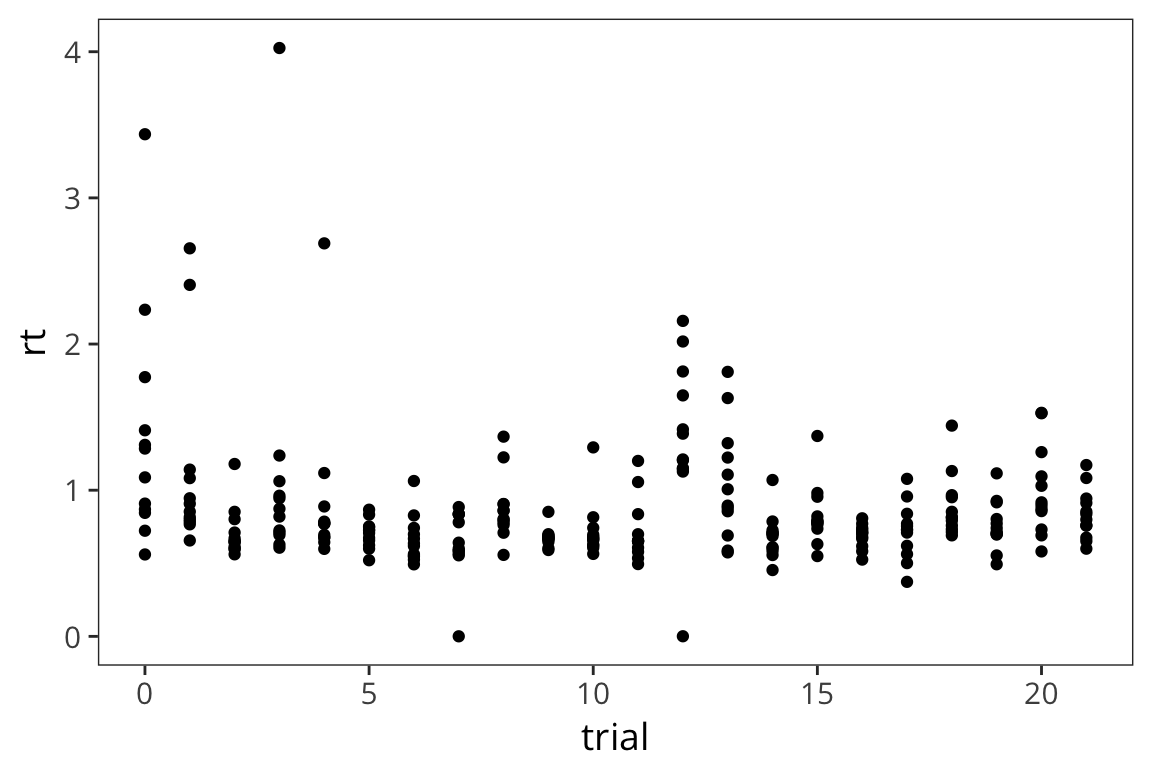
This is a fair number of data points so it’s a little difficult to see what’s going on. It might be useful to also show what the average across participants looks like at every timepoint. We can just add another layer to this same graph with the +, in this case we’ll use geom_line().
rt_by_trials <- rt_data %>%
group_by(trial) %>%
summarise(mean_rt = mean(rt))
ggplot() +
geom_point(data = rt_data, aes(x = trial, y = rt), alpha = 0.2) +
geom_line(data = rt_by_trials, aes(x = trial, y = mean_rt), color = "blue")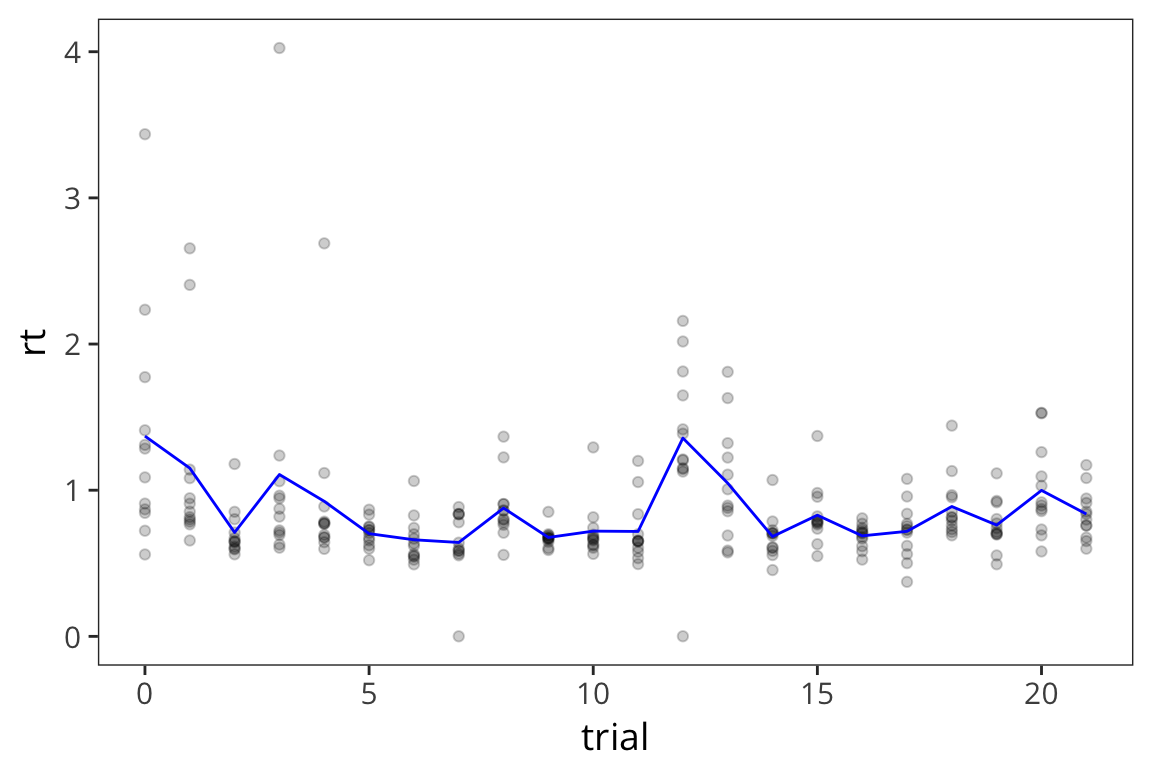
Because there were so many points and it was difficult to see, I made each point less opaque using
alpha = 0.2as an argument forgeom_point()and I made the line connecting averages stand out by making it blue withcolor = "blue"as an argument togeom_line()Note that I’m plotting 2 different datasets withing the same graph and this is easy to do because you can define data separately for each geom.
What can we learn from this scatterplot + line graph?
Is anything different happening in the first few trials? Does there seem to be a trend over the course of the trials?