Adam H Marblestone
Email: adam.h.marblestone [at] gmail
Publications: google scholar
Patents: google patents
CV: PDF

Implosion fabrication
with Dan Oran*, Sam Rodriques*, Rui Gao, Shoh Asano, Mark Scott, Fei Chen, Paul Tillberg and Ed Boyden

Lithographic nanofabrication is often limited to successive fabrication of two-dimensional (2D) layers. We present a strategy for the direct assembly of 3D nanomaterials consisting of metals, semiconductors, and biomolecules arranged in virtually any 3D geometry. We used hydrogels as scaffolds for volumetric deposition of materials at defined points in space. We then optically patterned these scaffolds in three dimensions, attached one or more functional materials, and then shrank and dehydrated them in a controlled way to achieve nanoscale feature sizes in a solid substrate. We demonstrate that our process, Implosion Fabrication (ImpFab), can directly write highly conductive, 3D silver nanostructures within an acrylic scaffold via volumetric silver deposition. Using ImpFab, we achieve resolutions in the tens of nanometers and complex, non–self-supporting 3D geometries of interest for optical metamaterials.
See also: nm2cm concept below
News and views by Long and Williams: HTML PDF
Patent: HTML
Protein Sequencing via Single-Molecule Kinetics
with Sam Rodriques and Ed Boyden
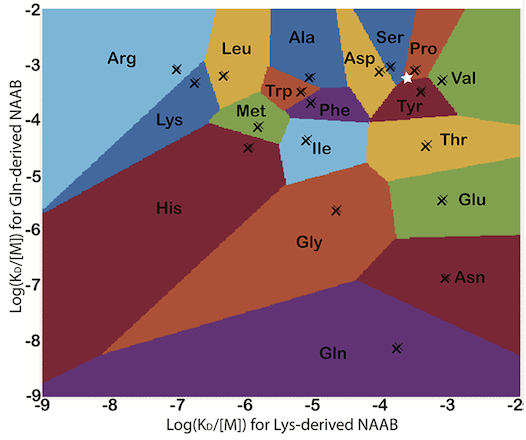
Next-gen DNA sequencing has revolutionized biology. But what if you could sequence individual proteins? Here we propose, and mathematically evaluate, a concept of how to do that. Specifically, we propose to measure the kinetics of diverse but weak N-terminal binding events in the time domain during Edman degradation to provide a unique profile of each amino acid along the protein chain, with readout via, for example, silicon single photon detectors arrayed on the chip on which the proteins are immobilized.
Paper: HTML
Preprint (June 2018): HTML
Rapid prototyping of 3D DNA-origami shapes with caDNAno
with Shawn Douglas and William Shih
Software: www.cadnano.org
Publication: PDF
Repositories of open caDNAno files: OpenWetWare and cadnano.org

I helped to write the first version of the open-source software CADnano for design of three-dimensional scaffolded DNA origami nanostructures. Shawn Douglas led the project and has since led the development of improved versions in collaboration with AutoDesk, Digizyme, the Wyss Institute and other groups. This is now the main design software used by the structural DNA nanotechnology field.
Exponential quantum enhancement for distributed addition with local nonlinearity
with Michel Devoret
Publication: PDF
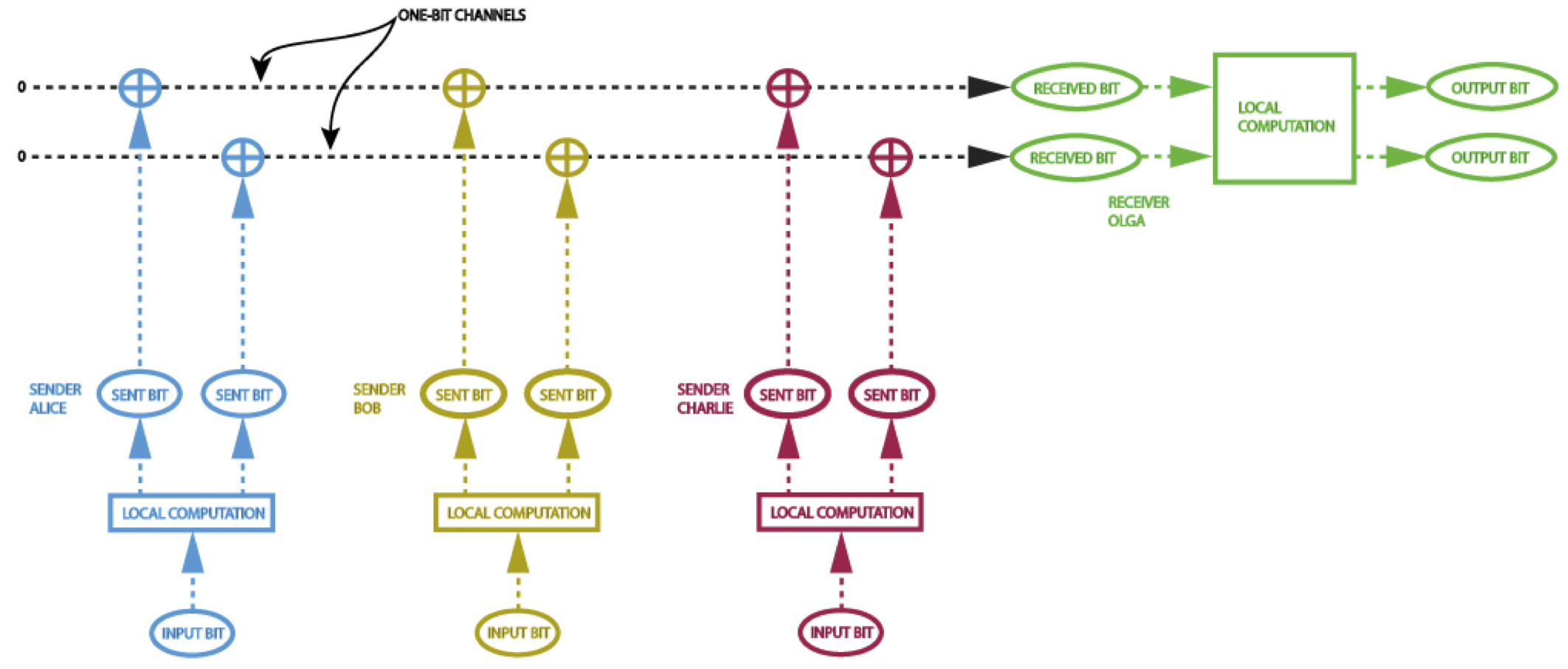
Using quantum entanglement, it is theoretically possible to perform so-called "pseudo-telepathy": groups of separated, non-communicating individuals can perform collective tasks which would, in a non-quantum universe, absolutely require them to communicate. To do so, they must establish quantum entanglement beforehand: the prior entanglement serves as a kind of substitute for later communication. Intrigued by limited examples of such quantum pseudo-telepathy schemes, I wondered whether quantum mechanics could allow complex computations (involving chains of non-linear logic operations) to be performed using less communication than would be required classically. Michel Devoret and I showed that this was true: the non-linear logic involved in performing a distributed binary addition operation can be done using exponentially less communication than would normally be necessary, provided the parties involved share prior entanglement.
nm2cm integration
with George Church, Robert Barish and others
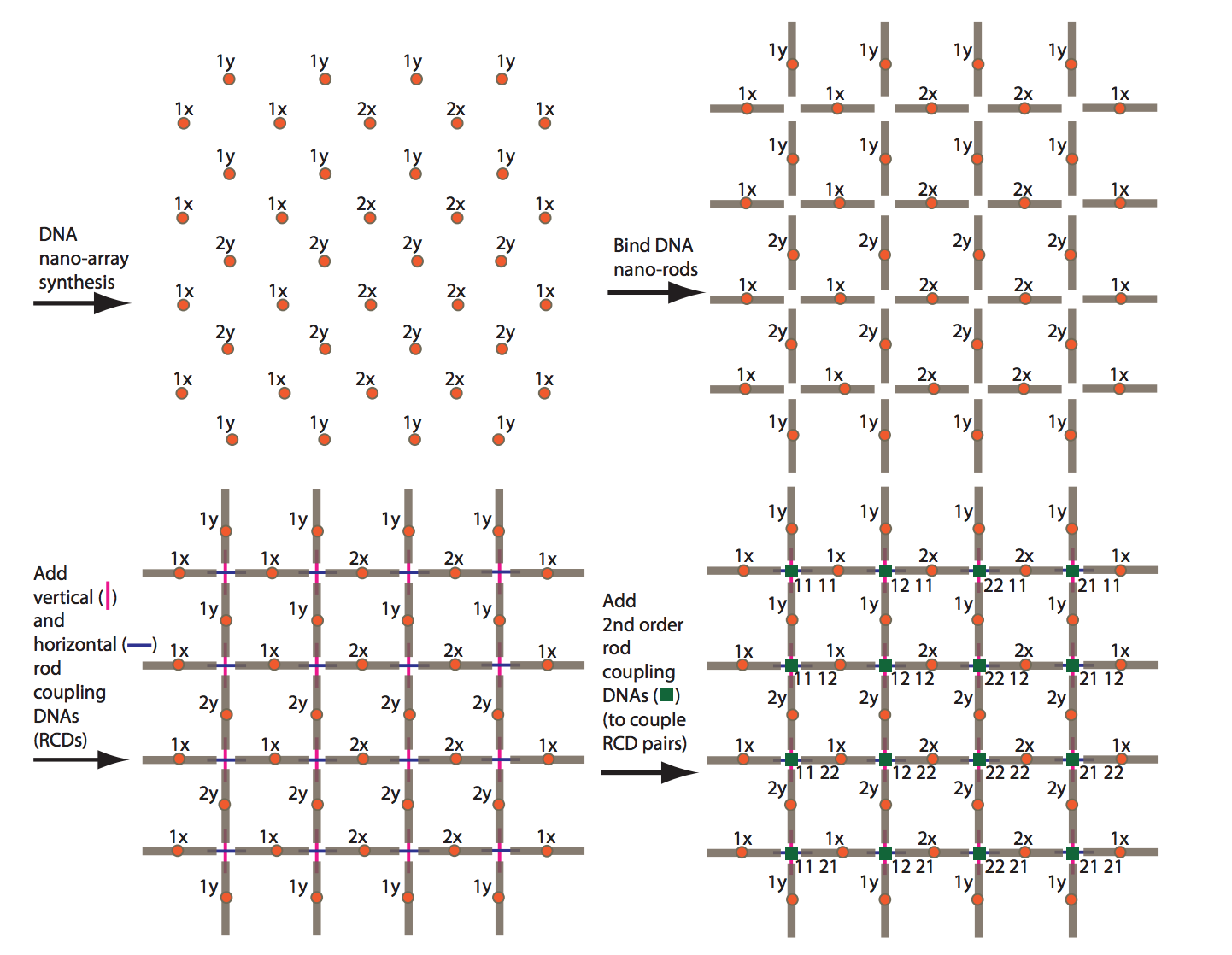
No integrated architecture has yet been proposed which fully specifies how to produce structures with a) overall sizes on the scale of today’s computer chips (centimeters), b) addressable features on the 10 nm scale, and c) the ability to attach a wide range of discrete molecular components at customizable locations. We are investigating schemes for nanometer-to-centimeter fabrication integration via top-down organization of DNA nanorods using DNA hybridization interactions, as well as other architectures.
Thesis excerpt: PDF
Talk slides: PDF
Molecular 3D printing
with Eric Drexler and Shahar Avin
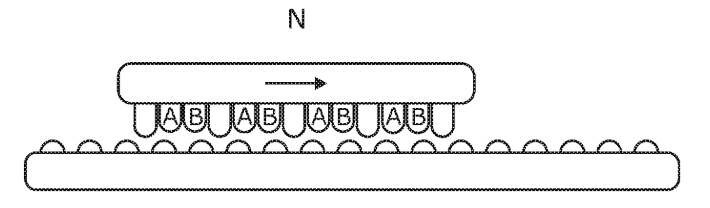
We organized a workshop at the University of Cambridge to explore molecular additive manufacturing: 3D printing with lego-like macro-molecular building blocks, sequential solid-phase and self-assembly, and dynamic nanoscale structural frameworks and motors based on DNA or protein origami.
OpenPhil writeup: PDF
Brainstorm on artificial ribosome programs: video
Unpublished bottleneck analysis of the positional chemistry field.
Seeing a single strand of DNA
with Mingjie Dai, Ralf Jungmann and Peng Yin
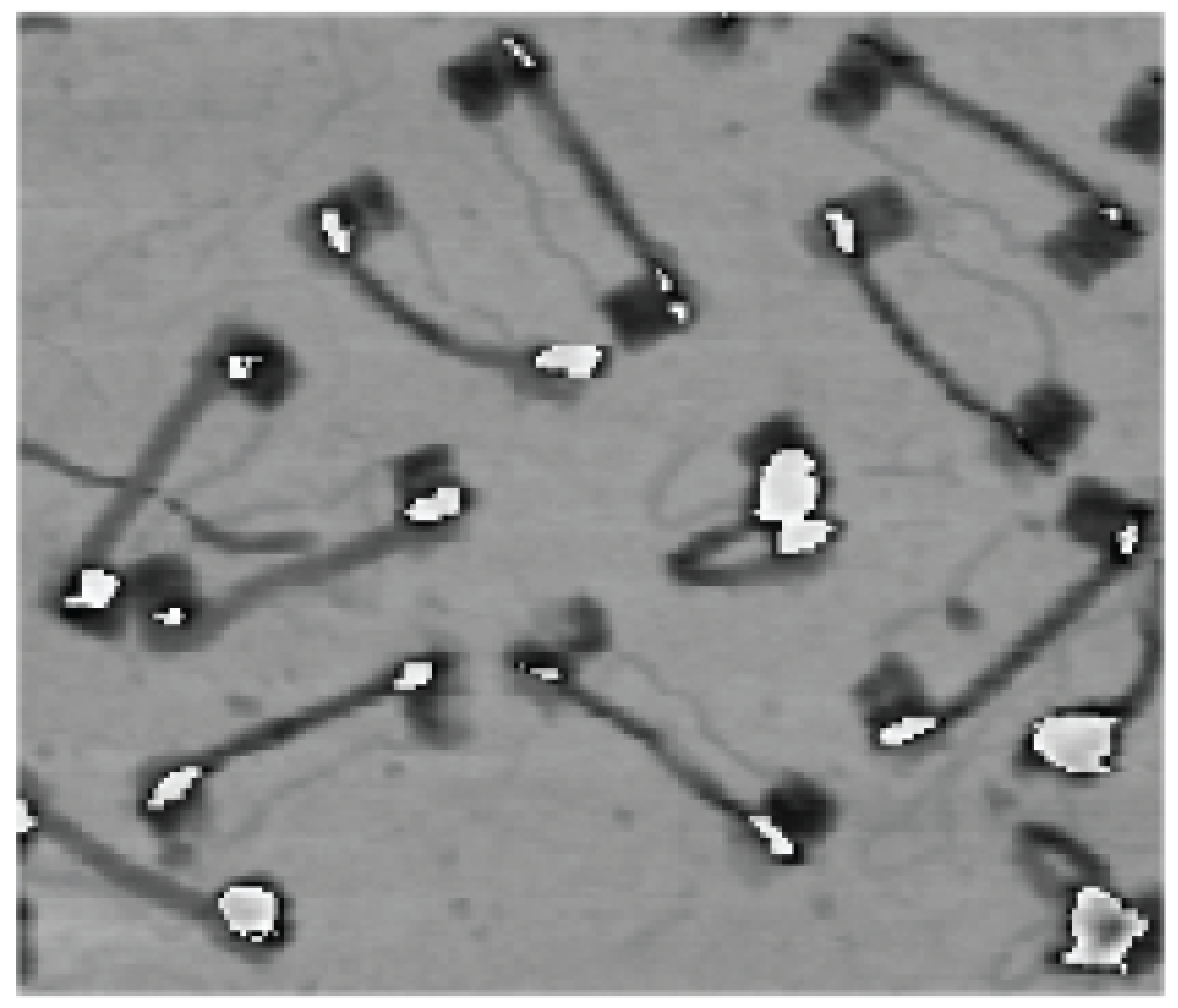
In this project, we made a nanoscale "tightrope" out of a single double helix of DNA.
Thesis excerpt: PDF
I helped to design a new type of amplifier for microwave-frequency electronic signals. The device operates in a regime where macroscopic electrical quantities like voltage and current exhibit "spooky" quantum behavior.