Adam H Marblestone
Email: adam.h.marblestone [at] gmail
Publications: google scholar
Patents: google patents
CV: PDF

Rosetta Brain: optical, molecularly-annotated connectomics
with Edward S Boyden, Richie Kohman, Samuel G Rodriques, Bobae An, Anthony M Zador, George M Church, Young-Gyu Yoon, Alex Vaughan, Reza Kalhor, Ruihan Zhang, Daniel Goodwin, Andrew G Xue and many collaborators
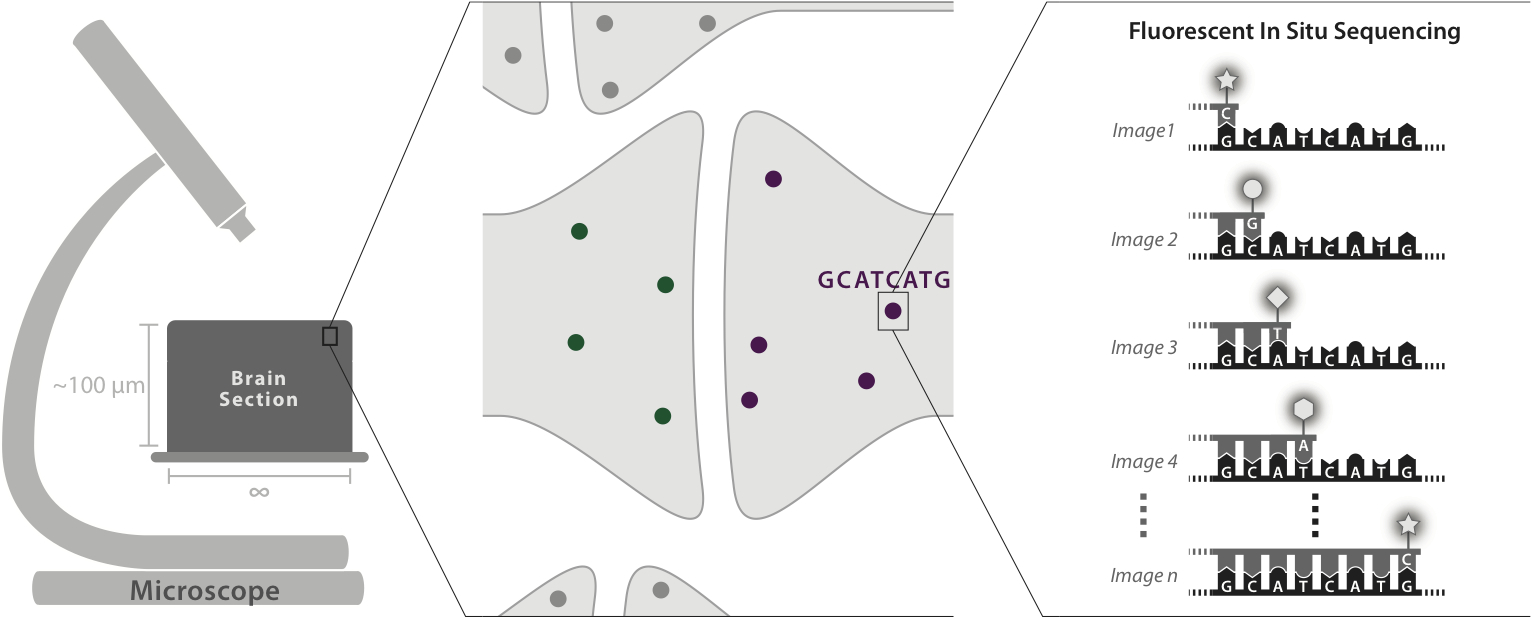
An optical approach to brain circuit mapping should enable two key capabilities:
1) High-throughput, robust neuron reconstruction over large scales, including entire mammalian brains -- through the use of molecular barcodes to uniquely tag each neuron with a distinguishable color combination
2) "Molecular annotation" of synaptic connection types, ion channel distributions within neurons, and other key properties -- by applying multiplexed optical readout to endogenous molecules.
Through the combination of expansion microscopy and in-situ molecular multiplexing, we can effectively achieve a kind of "arbitrary resolution (down to perhaps 10 nanometers), infinite color microscopy (up to perhaps 4 to the 20th power = trillions of distinct labels)" of fixed tissue. We hope that this will revolutionize connectomics, as well as other areas of biological tissue mapping.
Experimental integration of expansion microscopy and in-situ sequencing (ExSEQ): bioRxiv
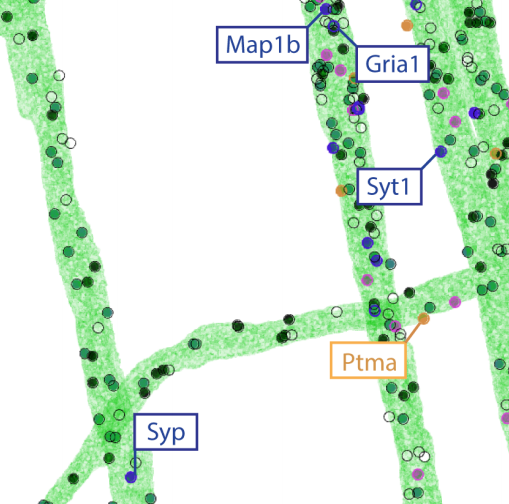
Experimental integration of ExSEQ and Zador barcodes: bioRxiv
Slides on these ideas (presented at Argonne National Lab): PDF
Computational paper on cell barcodes and 20x expansion microscopy for optical connectomic reconstruction: Frontiers
Expansion microscopy of lipid membranes: preprint
Early Rosetta Brain preprint on principles and goals of in-situ barcode connectomics: ArXiv
Early calculations outlining scalability considerations for connectomics approaches: bioRxiv
A simulation of FISSEQ-BOINC connectomics (showing one FISSEQ base plus morphological markers at 4x expansion): avi video
news: Some of our recent results on in-situ neuronal barcoding are featured in this article about the IARPA MICRONS project. Here is an image from that article showing in-situ sequencing of RNA barcodes in expanded mouse brain tissue -- the red and green represent two different RNA letters: 
news: The Rosetta Brain has been featured in an excellent popular book, The Future of the Brain, edited by Gary Marcus and Jeremy Freeman. Highly recommended!
news: The Rosetta Brain has been featured in a proposal for a National Network of Neurotechnology Centers, as a possible approach for molecularly annotated optical connectomics.
news: FISSEQ-BOINC connectomics approaches were a part of the IARPA MICRONS program to derive new machine learning algorithms by studying mesoscale cortical circuitry. I was MIT's PI on the project. Science picked up the story here.
news: From Dawen Cai's lab, BrainBow multiplexing plus Expansion Microscopy for optical connectomic mapping
Physical Principles for Scalable Neural Recording
with Bradley M. Zamft*, Yael G. Maguire, Mikhail G. Shapiro, Ted Cybulski, Joshua I. Glaser, P. Benjamin Stranges, Reza Kalhor, David A. Dalrymple, Dongjin Seo, Elad Alon, Michel M. Maharbiz, Jose Carmena, Jan Rabaey, Edward S. Boyden**, George M. Church** and Konrad P. Kording**

Simultaneously measuring the activities of all neurons in a mammalian brain at millisecond resolution is a challenge beyond the limits of existing techniques in neuroscience. Entirely new approaches may be required, motivating an analysis of the fundamental physical constraints on the problem. We outline the physical principles governing brain activity mapping using optical, electrical, magnetic resonance, and molecular modalities of neural recording.
Publisher link: Front. in Comp. Neurosci.
Preprint: ArXiv
Commentary in Nature Physics.
news: A group of collaborators has received an NIH BRAIN grant on interfacing with the brain via the bloodstream.
news: The Neurotechnology Architecting Network was featured by the White House as part of the BRAIN initiative. I got to give a presentation there on photonics technologies in neuroscience.
news: A related paper on "Spatial Information in Large-Scale Neural Recordings" has been published, studying the statistics of large-scale neural recordings.
news: In late 2016, several collaborators on this work including myself are tapped by the neurotechnology company Kernel to help develop the next generation of scalable human brain interfaces (and a few of the same also helped with initial ideation for Neuralink around the same time). See my EMTech talk about the approach I led as Kernel's Chief Strategy Officer, leading to their current focus on novel forms of non-invasive brain interfacing. We concluded that non-invasive brain interfacing had more physical headroom (no pun intended) to grow than was commonly appreciated.  DARPA concluded similarly about a year later. In early 2020, Kernel came out of stealth mode and announced its noninvasive hardware and neuroscience as a service programs. In mid 2020, Kernel raised $53 million in VC funding from leading investors towards these programs.
DARPA concluded similarly about a year later. In early 2020, Kernel came out of stealth mode and announced its noninvasive hardware and neuroscience as a service programs. In mid 2020, Kernel raised $53 million in VC funding from leading investors towards these programs.
Neural Recording via Optical Reflectometry
with Sam Rodriques, Max Mankin, Lowell Wood and Ed Boyden
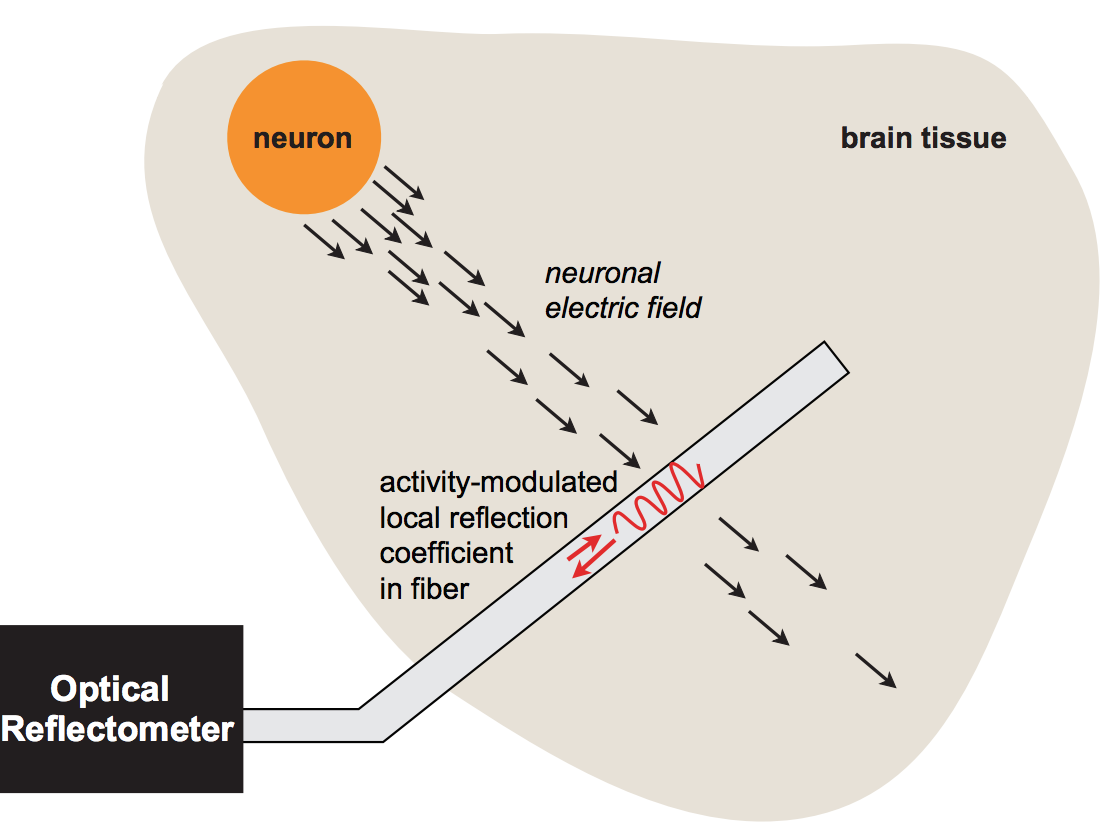
We introduce a fiber-optic architecture for neural recording without contrast agents, and study its properties theoretically. Our sensor design is inspired by electrooptic modulators. A key concept of the design is the ability to create an "intensified" electric field inside an optical waveguide by applying the extracellular voltage from a neural spike over a nanoscopic distance.
Journal of Biomedical Optics paper: html
Preprint: ArXiv
Towards an integration of neuroscience and deep learning
with Greg Wayne, Ken Hayworth and Konrad Kording
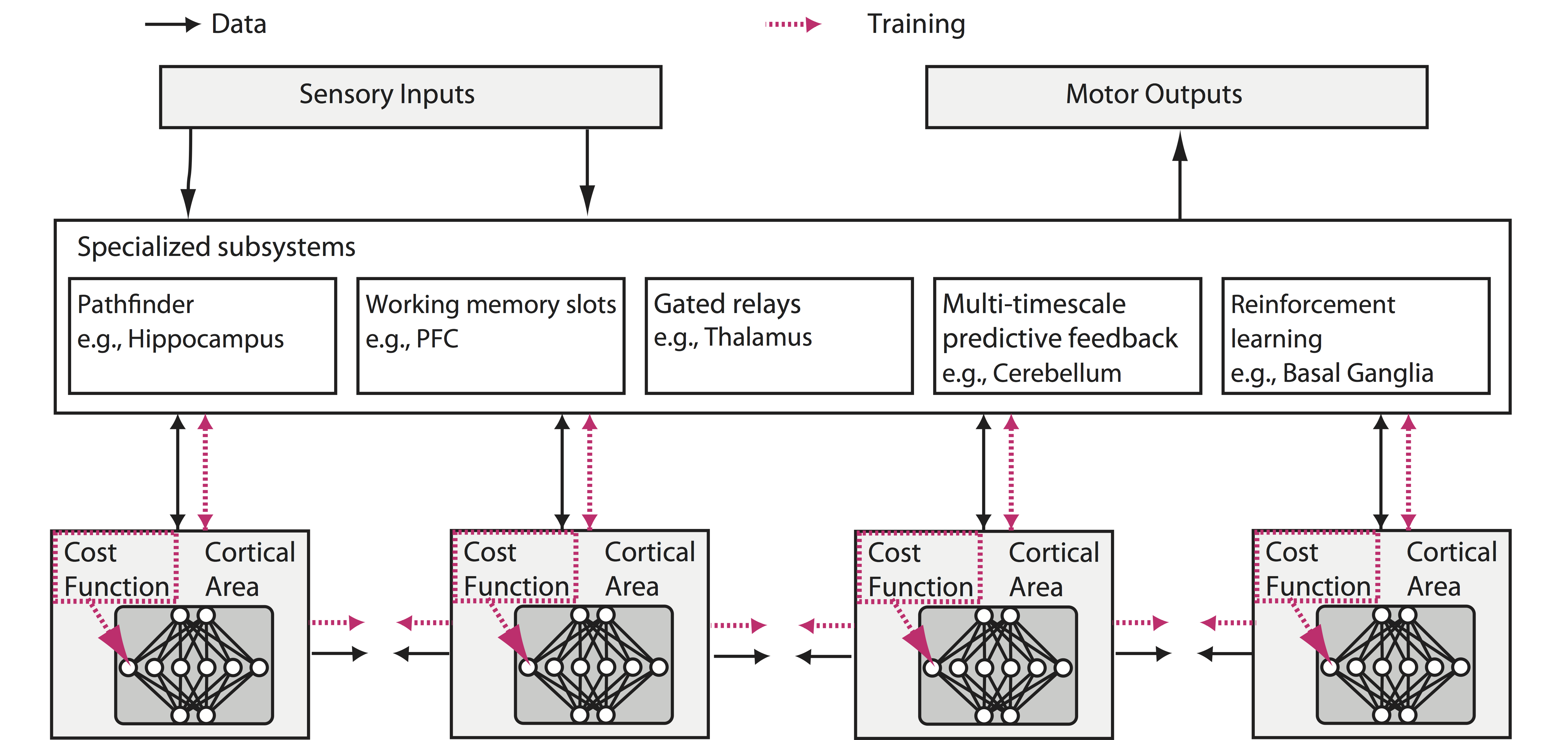
Neuroscience has focused on the detailed implementation of computation, studying neural codes, dynamics and circuits. In machine learning, however, artificial neural networks tend to eschew precisely designed codes, dynamics or circuits in favor of brute force optimization of a cost function, often using simple and relatively uniform initial architectures. Two recent developments have emerged within machine learning that create an opportunity to connect these seemingly divergent perspectives. First, structured architectures are used, including dedicated systems for attention, recursion and various forms of short- and long-term memory storage. Second, cost functions and training procedures have become more complex and are varied across layers and over time. Here we think about the brain in terms of these ideas. We hypothesize that (1) the brain optimizes cost functions, (2) these cost functions are diverse and differ across brain locations and over development, and (3) optimization operates within a pre-structured architecture matched to the computational problems posed by behavior. Such a heterogeneously optimized system, enabled by a series of interacting cost functions, serves to make learning data-efficient and precisely targeted to the needs of the organism. We suggest directions by which neuroscience could seek to refine and test these hypotheses.
Paper: BioRxiv
Slides: PDF
The Atoms of Neural Computation
with Gary Marcus and Tom Dean

The goal of this project is to catalyze the characterization of an integrative taxonomy and phylogeny of neural computations. A focus is on exploring the potential neural underpinnings of "variable binding", a problem of relevance to modern artificial intelligence.
Short essay: PDF
Publisher link: html
FAQ: ArXiv
Summary in NY Times Sunday Review.
These analyses led the Kavli Foundation to sponsor a series of workshops organized by Gary, Tomaso Poggio and myself. Here are the questions that we asked participants.
Thalamic relays, neural attractors and variable binding
work led by Ken Hayworth
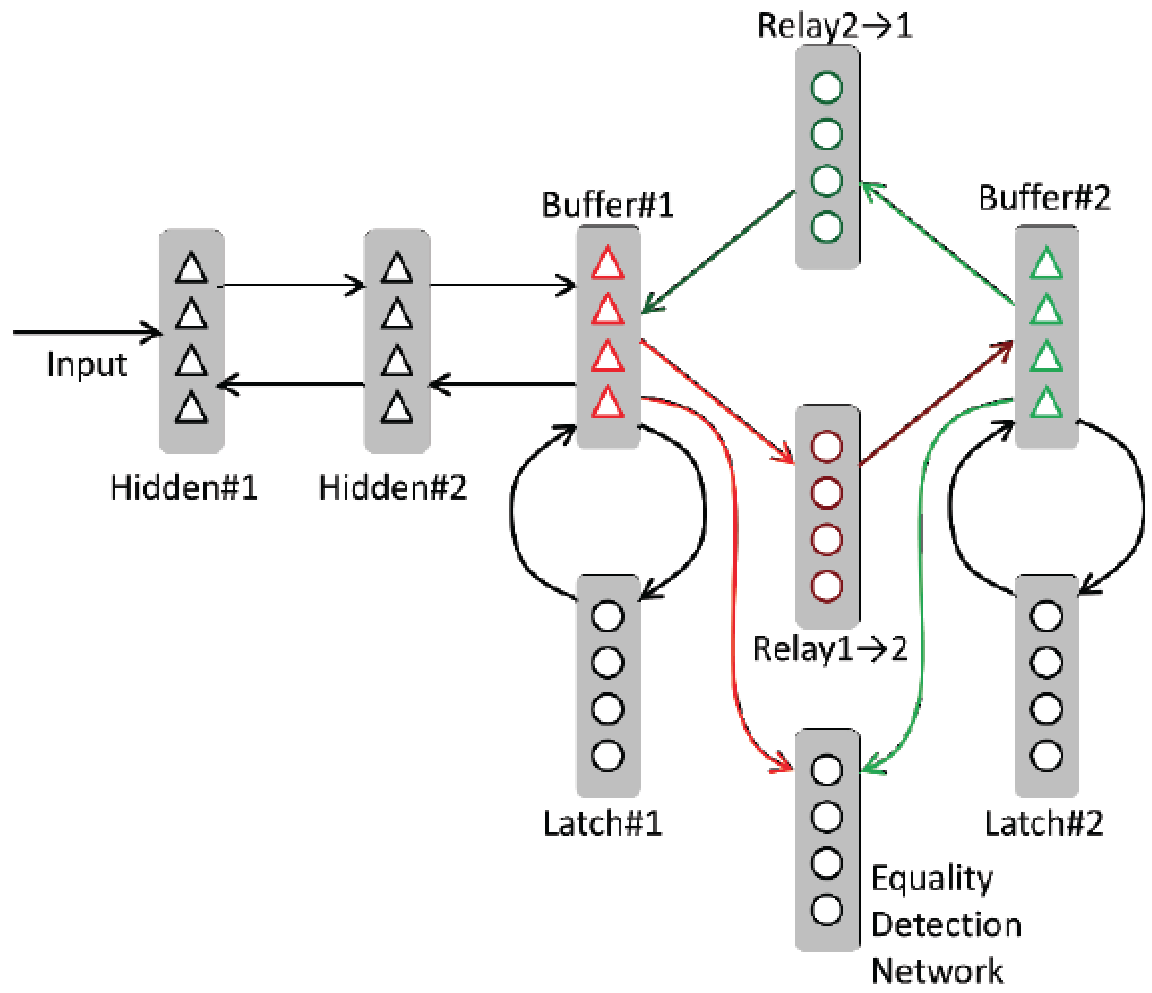
Preprint: bioRxiv
Slides on this: PDF
Lecture: Slides and Video
The thalamus appears to be involved in the flexible routing of information among cortical areas, yet the computational implications of such routing are only beginning to be explored. Here we create a connectionist model of how selectively gated cortico-thalamo-cortical relays could underpin both symbolic (e.g., "variable binding"), and sub-symbolic computations.
Molecular Tickertapes
with Brad Zamft*, Konrad Kording, Daniel Schmidt, Daniel Martin-Alarcon, Keith Tyo, Ed Boyden and George Church
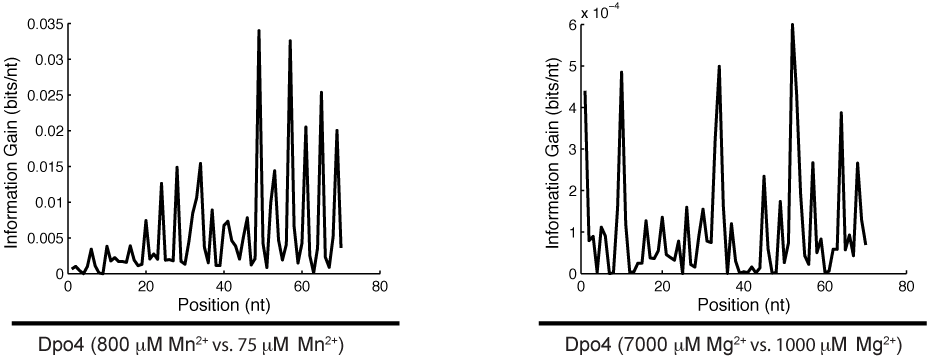
High-throughput recording of signals embedded within inaccessible micro-environments is a technological challenge. The ideal recording device would be a nanoscale machine capable of quantitatively transducing a wide range of variables into a molecular recording medium suitable for long-term storage and facile readout in the form of digital data. We have recently proposed such a device, in which cation concentrations modulate the misincorporation rate of a DNA polymerase (DNAP) on a known template, allowing DNA sequences to encode information about the local cation concentration. In this work we quantify the cation sensitivity of DNAP misincorporation rates, making possible the indirect readout of cation concentration by DNA sequencing.
Publication on using next-generation sequencing to measure ion-dependent DNA polymerase error rates: HTML
Publication on improved next-generation sequencing based polymerase error mapping using nucleotide imbalances: HTML
Note: this project is still at a very preliminary stage of experimentation!
news: We have published a theoretical paper (Joshua I. Glaser et al, PLoS Computational Biology) on the statistical inverse problems associated with molecular ticker-tapes, and how they constrain the synthetic biology implementation challenges.
news: We have received a Transformative Research grant from the NIH to further investigate the recording of dynamic neuronal activities into biopolymer data storage media! The full list of 2013 awardees is here.
Commentary in Nature.