Adam H Marblestone
Email: adam.h.marblestone [at] gmail
Publications: google scholar
Patents: google patents
CV: PDF

Some computational neuroscience code notebooks

C. elegans network simulation Github (based on Kunert et al and OpenWorm) (see slides)
Expander graph Hopfield network Github (based on Chaudhuri and Fiete)
Cognitive Integration
with Joscha Bach and Manushaqe Muco

After decades of separation, it is time to re-integrate the sciences of cognition into a cohesive understanding. Cognitive Architectures, autonomous learning, types and modes of representation and constraints from neuroscience offer new ways of looking at mind and intelligence. The Cognitive Integration class at the MIT Media Lab consists of a theoretical part, focused on concepts and discussion, and practical hands-on sessions, with the goal of implementing AI models in an agent based paradigm.
Revolutionary Ventures
with Ed Boyden, Joost Bonsen, Desiree Dudley and Joe Jacobson
The Revolutionary Ventures class evolved from Ed and Joost's Neurotechnology Ventures. It covers envisioning and building ideas and organizations to accelerate engineering revolutions. Focuses on emerging technology domains, such as neurotechnology, imaging, cryotechnology, gerontechnology, and bio-and-nano fabrication. Draws on historical examples as well as live case studies of existing or emerging organizations, including labs, institutes, startups, and companies. Goals range from accelerating basic science to developing transformative products or therapeutics. Each class is devoted to a specific area, often with invited speakers, exploring issues from the deeply technical through the strategic. Individually or in small groups, students prototype new ventures aimed at inventing and deploying revolutionary technologies.
C. elegans optogenetics for MIT's Neurotechnology In Action lab
with Ed Boyden, Desiree Dudley, Demian Park, Nikhil Bhatla, Dengke Ma, Jay Yu and Maxine Jonas
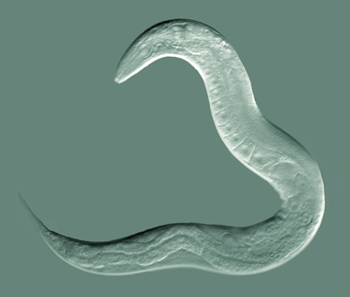
We created a simple demo lab for demonstrating optogenetic control of C. elegans behavior using cheap LEDs and Arduino's. The students make the nematode freeze its motion, curl up or change direction in response to blue light.
Strains and setup: PDF
Arduino-driven lamp: mov
Bio-molecular Engineering and Synthetic Biology
with William Shih, Peng Yin and Pam Silver

I helped create Harvard's class on DNA nanotechnology and molecular programming and was its first teaching assistant. Graduate level seminar focusing on in-silico design pipelines for nucleic acid- and protein-based molecular machinery and programmable molecular systems. Students are mentored to produce substantial midterm and final group design projects.
Expii Physics Knowledge Graph
with Po-Shen Loh and the Expii Team
I was part of the team that created the first Physics knowledge graphs for Expii.
After the company Halycon Molecular folded over, I supervised Andrew Payne in the Church lab to write a publication reproducing, characterizing and documenting their DNA manipulation technology.
Nanoscale containers and other nanoscale structures
with Nick Perkons, Sherrie Wang, Evan Wu, Wei Sun, Tom Schaus, William Shih and Peng Yin

I mentored a team on a project to design and construct molecular containers.
Designs: Wiki
news: Our team won 3rd prize overall in the BioMod 2011 international competition!
L-DNA templating
with Wesley Chen, Ian Choi, In-Young Cho, Valentina Lyau, Jacob Pritt, Mingjie Dai, Ralf Jungmann, William Shih, Jagesh Shah and Peng Yin

It is now possible to create fully-addressable nanostructures from hundreds of unique synthetic DNA strands. Such structures, being made of biologically-natural D-DNA, are easily degraded by nucleases, posing a potential problem for in-vivo applications such as drug delivery. The oppositely-chiral L-DNA cannot be so degraded due to the inability of chiral nucleases to recognize and attack its mirror-image structure. Unfortunately, L-DNA is currently much more expensive than D-DNA. Here my students demonstrate a method for inexpensively creating L-DNA structures with controlled shapes: a repetitive L-DNA structure is templated on an inexpensive, all-unique D-DNA layer. The D-DNA layer forms first, exposing D-DNA handles at defined locations. Subsequently, alternating rows of identical L-DNA/D-DNA chimerical strands are added through hybridization of complementary handles.
Designs: Wiki
news: Our team won Best Wiki in the BioMod 2012 international competition!
Other courses I like:
- McBride on physical organic chemistry and quantum mechanics thereof
- Phillips on physical biology of the cell
- David MacKay on information theory
- De Freitas course on deep learning at Oxford
- Silver course on reinforcement learning
- How to Make (Almost) Anything
- The Physics of Information Technology
- How to Grow (Almost) Anything
- The Nature of Mathematical Modeling
- Nonlinear Dynamics
- The Human Intelligence Enterprise
- Biomolecular Engineering and Synthetic Biology
- Probabilistic Agent Learning
- Computational Models of the Neocortex
- How to build a brain from scratch
- All the key ideas of linear algebra in one lesson (Gilbert Strang)
- Hamprecht on machine learning
- Graphical models course at Stanford
- Seminar on Towards an Integration of Deep Learning and Neuroscience
